For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNLILTAHGTRRPSGVAMIADIAAQVSALVDRTVQVAFVDVLGPSPSEVLSALSCRPAIVVPAFLSRGYH VRTDLPAHVAASAHPHVTVTPALGPCREIAQIVTQQLVESGWRPGDSVILAAAGASDRRARADLHTTRTL VSELTGSWVDMGFAGTGGPDVRTAVQRARDRAEANRGARRVVVASFLLAEGLFQERLRASGADVVTRPLG THPGLAQLVANRFRSAVARQQRLHRWHGTPTPVTLDL
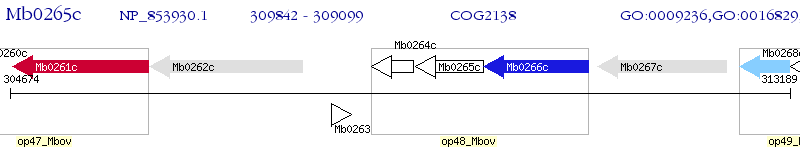
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0265c | - | - | 100% (247) | hypothetical protein Mb0265c |
| M. bovis AF2122 / 97 | Mb2414 | - | 3e-22 | 33.19% (235) | hypothetical protein Mb2414 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0381 | - | 8e-69 | 58.85% (226) | cobalamin (vitamin B12) biosynthesis CbiX protein |
| M. tuberculosis H37Rv | Rv0259c | - | 1e-138 | 99.60% (247) | hypothetical protein Rv0259c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3519c | - | 1e-53 | 49.13% (230) | hypothetical protein MAB_3519c |
| M. marinum M | MMAR_0519 | - | 2e-86 | 72.10% (233) | hypothetical protein MMAR_0519 |
| M. avium 104 | MAV_4901 | - | 4e-80 | 68.12% (229) | sirohydrochlorin cobaltochelatase, putative |
| M. smegmatis MC2 155 | MSMEG_0431 | - | 5e-73 | 59.47% (227) | secreted protein |
| M. thermoresistible (build 8) | TH_1445 | - | 1e-18 | 32.48% (234) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1182 | - | 6e-86 | 71.67% (233) | hypothetical protein MUL_1182 |
| M. vanbaalenii PYR-1 | Mvan_0300 | - | 2e-70 | 57.74% (239) | cobalamin (vitamin B12) biosynthesis CbiX protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0381|M.gilvum_PYR-GCK ---------------MSLVLVAHGTRKQRGVSLIGDLSARVS-AVLDRPV
Mvan_0300|M.vanbaalenii_PYR-1 ---------------MSLLLVAHGTRKQRGVSLIGDIAAQVS-TALGRPV
MSMEG_0431|M.smegmatis_MC2_155 ---------------MSLLLVAHGTRKPGGVAMIADLAQRVA-EQLERQV
Mb0265c|M.bovis_AF2122/97 ---------------MNLILTAHGTRRPSGVAMIADIAAQVS-ALVDRTV
Rv0259c|M.tuberculosis_H37Rv ---------------MNLILTAHGTRRPSGVAMIADIAAQVS-ALVDRTV
MMAR_0519|M.marinum_M ---------------MHLILTAHGTRRPAGVAMIGDLAARVS-ALLKQTV
MUL_1182|M.ulcerans_Agy99 ---------------MHLILTAHGTRRPAGVAMIGDLAARVS-ALLKQTV
MAV_4901|M.avium_104 ---------------MALILVAHGTRRPGGVAMIEGLAAQVS-TLVGGRV
MAB_3519c|M.abscessus_ATCC_199 --------------MAEFVLVAHGTRSAAGVENIAALAEAVS-RRVG-SV
TH_1445|M.thermoresistible__bu VTRALAPAPASVAGPPALILTAHGSADPRSAANAHAIAGHLRRLAPDVEV
::*.***: .. :: : *
Mflv_0381|M.gilvum_PYR-GCK RVAFVDVLGPTPSELLR-----EEFHRPTVLVPAFLSRGYHVSRDIPSHV
Mvan_0300|M.vanbaalenii_PYR-1 RVAFVDVLGPTPAELLG-----ATRHEPTVLVPAFLARGYHVSSDIPAHV
MSMEG_0431|M.smegmatis_MC2_155 QVAFVDVLGPTPRDVLDT----MAADRPVVLVPAFLAAGYHVRVDVPAHV
Mb0265c|M.bovis_AF2122/97 QVAFVDVLGPSPSEVLS-----ALSCRPAIVVPAFLSRGYHVRTDLPAHV
Rv0259c|M.tuberculosis_H37Rv QVAFVDVLGPSPSEVLS-----ALSCRPAIVVPAFLSRGYHVRTDLPAHV
MMAR_0519|M.marinum_M QVAFVDVLGPTPSEVLGRPAPGRGSAQSAIVVPAFLSRGYHVRIDLPAHV
MUL_1182|M.ulcerans_Agy99 QVAFVDVLGPTPSEVLGRPAPGRGSAQSAIVVPAFLSRGYHVRIDLPAHV
MAV_4901|M.avium_104 EVAFVDVVGPTPSEVLA---AARAAGRPAIVVPAFLSRGYHVRADLPAHV
MAB_3519c|M.abscessus_ATCC_199 RTAFVDVLGPTPSEVLS------TIDGPAVLLPAFLASGYHVHHDIPEHV
TH_1445|M.thermoresistible__bu RVAFCEQNTPNLADVLA------QMRTGAVVVPLLLARAYHARVDIPELI
..** : *. ::* .:::* :*: .**. *:* :
Mflv_0381|M.gilvum_PYR-GCK EASGHREVTVTRALGPDPALIRVLVDRLVESGCHPEDS---VILAAAGTS
Mvan_0300|M.vanbaalenii_PYR-1 AASGHDDVLVTRALGPDPALVRVLVDRLIESGWRPNDS---VILAAAGTS
MSMEG_0431|M.smegmatis_MC2_155 SASGHPLVTVTPPLGPCHGTVRVLADRLVESGWQPGDA---VILAAAGTS
Mb0265c|M.bovis_AF2122/97 AASAHPHVTVTPALGPCREIAQIVTQQLVESGWRPGDS---VILAAAGAS
Rv0259c|M.tuberculosis_H37Rv AASAHPHVTVTPALGPCREIAQIVTQQLVESGWRPGDS---VILAAAGAS
MMAR_0519|M.marinum_M AASGHPDVTVTPALGPSREIARILIQQLLESGWRPDDS---VILAAAGTS
MUL_1182|M.ulcerans_Agy99 AASGHPDVTVTPALGPSREIARILIQQLLEFGWRPDDS---VILAAAGTS
MAV_4901|M.avium_104 ALSGHQNVTVTPALGPSGQIARIVGDQLLECGWRPNDS---VVLAAAGTS
MAB_3519c|M.abscessus_ATCC_199 ALSGHPQVAITQTLGPDPVLASVLAGRLRAAGWRRGDA---VVLAAAGSS
TH_1445|M.thermoresistible__bu AASG-ADVAQADVLGADPRLLHVMRGRLSALGVSRFDAGLGVVMVGVGSS
*. * : **. :: :* * *: *::...*:*
Mflv_0381|M.gilvum_PYR-GCK DPAAMRDLHTTAAALSA-TIGSRVELAFAATGEPRVSDAVAEVRR-----
Mvan_0300|M.vanbaalenii_PYR-1 DPAAMRDLHTTATWLSA-TIGARVELAFAATGEPRIADAVAGLRR-----
MSMEG_0431|M.smegmatis_MC2_155 DPYAQSDLRRTAAMLSA-VTGDRVELAYAATGAPSVADAVAAQRR-----
Mb0265c|M.bovis_AF2122/97 DRRARADLHTTRTLVSE-LTGSWVDMGFAGTGGPDVRTAVQRARDRAEAN
Rv0259c|M.tuberculosis_H37Rv DRRARADLHTTRTLVSE-LTGSWVDMGFAGTGGPDVRTAVQRARDRAEAN
MMAR_0519|M.marinum_M DRRALDDLRTARAFLCA-LTGSPVTLGFAATGRPDLRTAVRAARR-----
MUL_1182|M.ulcerans_Agy99 DRRALDDLRTARAFLCA-LTGSPVTLGFAATGRPDLRTAVRAARR-----
MAV_4901|M.avium_104 DDKARADLHTAATWLSA-LTGSRVTLGFAATGDPPLGEAVARARPHAR--
MAB_3519c|M.abscessus_ATCC_199 DARARHEVHTAASLLAR-HTG-PVRVGYIATGEPRVADVVREARA-----
TH_1445|M.thermoresistible__bu HADANARTAAVAQALGEGTRWTGVEVAFATGPAPSVPDAVARLRA-----
. * . : * :.: * : .* *
Mflv_0381|M.gilvum_PYR-GCK RGCRRVVVASYLLSEGLFQDRLR---DSGADLVTDPLGTHPGVTRLIASR
Mvan_0300|M.vanbaalenii_PYR-1 PG-KRVVVASYLLSEGLFQDRLR---DSGADVVTDPLGTHPGVARLIANR
MSMEG_0431|M.smegmatis_MC2_155 RSDRRVVVASYLLAEGLFQDRLR---EAGADLVTAPLGTHGGMVRLVANR
Mb0265c|M.bovis_AF2122/97 RGARRVVVASFLLAEGLFQERLR---ASGADVVTRPLGTHPGLAQLVANR
Rv0259c|M.tuberculosis_H37Rv RGARRVAVASFLLAEGLFQERLR---ASGADVVTRPLGTHPGLAQLVANR
MMAR_0519|M.marinum_M NGARRVVVVSYLLAEGLFQERLR---ACGADVVTPPLGNHPGLAELIATR
MUL_1182|M.ulcerans_Agy99 NGARRVVVVSYLLAEGLFQERLR---ACGADVVTPPLGNHPGLAELIATR
MAV_4901|M.avium_104 RNGGRVVVASYLLADGLFQQRLH---GCGADLVSAPLSTHPGLARLIANR
MAB_3519c|M.abscessus_ATCC_199 -TGRRVFIASYLLAQGLFQQRLA---ECGADGVAQPLGVHPQIVDLLVQR
TH_1445|M.thermoresistible__bu RGADRIAISPWFLAHGRITDRVAGFARGAGIPMAEPLGPHRLVAATVLDR
*: : .::*:.* : :*: .. :: **. * :. : *
Mflv_0381|M.gilvum_PYR-GCK FQRAGYDTPCYDTPAEIAFQQRNVEADSYGQCNIR-
Mvan_0300|M.vanbaalenii_PYR-1 FQRAG-----FDTAAEIAFQQRTFEYAPAGMQFRR-
MSMEG_0431|M.smegmatis_MC2_155 FRRAG----VLSLPAAA-------------------
Mb0265c|M.bovis_AF2122/97 FRSAV-----ARQQ----RLHRWHGTPTPVTLDL--
Rv0259c|M.tuberculosis_H37Rv FRSAV-----ARQQ----RLHRWHGTPTPVTLDL--
MMAR_0519|M.marinum_M FHSAI-----SAAVPTVPRSPMHTRAESHETQSIVG
MUL_1182|M.ulcerans_Agy99 FHSAI-----SAAVPTVPRSPMHTRAESHETQSIVG
MAV_4901|M.avium_104 FRRALPPVLAATARHASRRTGPHQRAHAPATRPVP-
MAB_3519c|M.abscessus_ATCC_199 FASVT-------------AASGELEAVADVA-----
TH_1445|M.thermoresistible__bu YEETV-------------------AAAALAA-----
: .