For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPARSVPRPRWVAPVRRVGRLAVWDRPERRSGIPALDGLRAIAVALVLASHGGIPGMGGGFIGVDAFFVL SGFLITSLLLDELGRTGRIDLSGFWIRRARRLLPALVLMVLTVSAARALFPDQALTGLRSDAIAAFLWTA NWRFVAQNTDYFTQGAPPSPLQHTWSLGVEEQYYVVWPLLLIGATLLLAARARRRCRRATVGGVRFAAFL IASLGTMASATAAVAFTSAATRDRIYFGTDTRAQALLIGSAAAALLVRDWPSLNRGWCLIRTRWGRRIAR LLPFVGLAGLAVTTHVATGSVGEFRHGLLIVVAGAAVIVVASVAMEQRGAVARILAWRPLVWLGTISYGV YLWHWPIFLALNGQRTGWSGPALFAARCAATVVLAGASWWLIEQPIRRWRPARVPLLPLAAATVASAAAV TMLVVPVGAGPGLREIGLPPGVSAVAAVSPSPPEASQPAPGPRDPNRPFTVSVFGDSIGWTLMHYLPPTP GFRFIDHTVIGCSLVRGTPYRYIGQTLEQRAECDGWPARWSAQVNRDQPDVALLIVGRWETVDRVNEGRW THIGDPTFDAYLNAELQRALSIVGSTGVRVMVTTVPYSRGGEKPDGRLYPEDQPERVNKWNAMLHNAISQ HSNVGMIDLNKKLCPDGVYTAKVDGIKVRSDGVHLTQEGVKWLIPWLEDSVRVAS
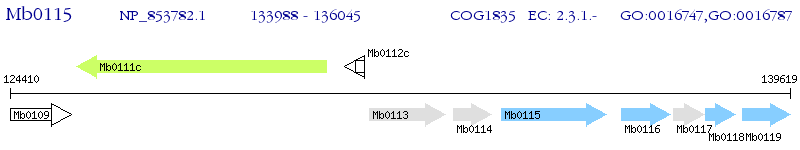
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0115 | - | - | 100% (685) | transmembrane acyltransferase |
| M. bovis AF2122 / 97 | Mb1592c | - | 6e-29 | 29.45% (421) | hypothetical protein Mb1592c |
| M. bovis AF2122 / 97 | Mb0233 | - | 1e-12 | 23.59% (407) | integral membrane acyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1859 | - | 0.0 | 61.69% (650) | acyltransferase 3 |
| M. tuberculosis H37Rv | Rv0111 | - | 0.0 | 100.00% (685) | transmembrane acyltransferase |
| M. leprae Br4923 | MLBr_02670 | - | 0.0 | 78.89% (668) | hypothetical protein MLBr_02670 |
| M. abscessus ATCC 19977 | MAB_2689 | - | 4e-32 | 31.85% (405) | acyltransferase |
| M. marinum M | MMAR_0312 | - | 0.0 | 82.39% (670) | transmembrane acyltransferase |
| M. avium 104 | MAV_5201 | - | 0.0 | 82.78% (668) | putative acyltransferase |
| M. smegmatis MC2 155 | MSMEG_5537 | - | 0.0 | 60.48% (673) | integral membrane protein |
| M. thermoresistible (build 8) | TH_1216 | - | 5e-83 | 67.34% (199) | POSSIBLE TRANSMEMBRANE ACYLTRANSFERASE |
| M. ulcerans Agy99 | MUL_4810 | - | 0.0 | 82.09% (670) | transmembrane acyltransferase |
| M. vanbaalenii PYR-1 | Mvan_4875 | - | 0.0 | 62.62% (650) | acyltransferase 3 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1859|M.gilvum_PYR-GCK -----------MTAVSAAPIRVGPRRYVGPGQ-SIPALDGVRAIAVALVL
Mvan_4875|M.vanbaalenii_PYR-1 -----------MTAVSAAPIRVGPRRYVGPGQ-AIPALDGVRAIAVALVL
MSMEG_5537|M.smegmatis_MC2_155 --------------MTAAPVRAAGRRRTLPARGEIPALDGIRAIAVLLVL
TH_1216|M.thermoresistible__bu --------------------------------------------------
Mb0115|M.bovis_AF2122/97 --MPARSVPRPRWVAPVRRVGRLAVWDRPERRSGIPALDGLRAIAVALVL
Rv0111|M.tuberculosis_H37Rv --MPARSVPRPRWVAPVRRVGRLAVWDRPERRSGIPALDGLRAIAVALVL
MMAR_0312|M.marinum_M ----------------MRRAGRTAIWDRAGRQRGIPALDGLRAVAVALVL
MUL_4810|M.ulcerans_Agy99 ----------------MRRAGRTAIWDRAGRQRGIPALDGLRAVAVALVL
MAV_5201|M.avium_104 ----------------MRRVGRLAIWDRPERRSGIPALDGLRAIAVALVL
MLBr_02670|M.leprae_Br4923 ----------------MRRVGLLRIWNQPERRNGIPALDGLRAIAVALVL
MAB_2689|M.abscessus_ATCC_1997 MFFVSATKPTKDPEVSPAAAMDATPKPKGDKAFYRYDLDGLRGIAIFLVA
Mflv_1859|M.gilvum_PYR-GCK ADHGGVPGLSGGFLGVDVFFVLSGFLITSLLLDEIGRTGR-VGLKGFWIR
Mvan_4875|M.vanbaalenii_PYR-1 ADHGGVPGLSGGFLGVDVFFVLSGFLITSLLLDEVGRTGR-IGLKGFWIR
MSMEG_5537|M.smegmatis_MC2_155 AGHAGVPGVAGGFIGVDIFFVLSGFLITSLLLDEFARTGR-VDLKGFWIR
TH_1216|M.thermoresistible__bu --------------------------------------------------
Mb0115|M.bovis_AF2122/97 ASHGGIPGMGGGFIGVDAFFVLSGFLITSLLLDELGRTGR-IDLSGFWIR
Rv0111|M.tuberculosis_H37Rv ASHGGIPGMGGGFIGVDAFFVLSGFLITSLLLDELGRTGR-IDLSGFWIR
MMAR_0312|M.marinum_M VGHGGVPGVGGGFIGVDIFFVLSGFLITSLLLDELGRSGR-IDLTGFWIR
MUL_4810|M.ulcerans_Agy99 VGHGGVPGVGGGFIGVDVFFVLSGFLITSLLLDELGRSGR-IDLTGFWIR
MAV_5201|M.avium_104 IGHGGIPGVSGGFIGVDVFFVLSGFLITSLLLDELGRTGR-IELTGFWIR
MLBr_02670|M.leprae_Br4923 TQHGGIPGLSGGFIGVDLFFVLSGFLITSLLLDELRHTGR-IDAINFWIR
MAB_2689|M.abscessus_ATCC_1997 VFHVWFGRVSG---GVDVFLTLSGFFYGSKLLRTATTQGASLNPVPVVKR
Mflv_1859|M.gilvum_PYR-GCK RARRLLPALVVMVLAVVAVRGLFSDES-VANLRDDAVATFFWMSNWIFVA
Mvan_4875|M.vanbaalenii_PYR-1 RARRLLPALLVMVLAVVALRELFSEES-TANLREDAVAAFFWMSNWVFVA
MSMEG_5537|M.smegmatis_MC2_155 RARRLLPALLVMTLAVVAARELFPPDA-VTSLRDDAVAAFFWVANWAFVA
TH_1216|M.thermoresistible__bu --------------------------------------------------
Mb0115|M.bovis_AF2122/97 RARRLLPALVLMVLTVSAARALFPDQA-LTGLRSDAIAAFLWTANWRFVA
Rv0111|M.tuberculosis_H37Rv RARRLLPALVLMVLTVSAARALFPDQA-LTGLRSDAIAAFLWTANWRFVA
MMAR_0312|M.marinum_M RARRLLPALVLMVLTVAAARELLPYQA-LTGLRNDALAAFLWMANWRFVA
MUL_4810|M.ulcerans_Agy99 RARRLLPALVLMVLTVAAARELLPYQA-LTGLRNDALAAFLWMANWRFVA
MAV_5201|M.avium_104 RARRLLPALVLMVLTVAAARQLLPERS-LTGLRDDAIAAFLWMANWRFVA
MLBr_02670|M.leprae_Br4923 RACRLLPALVLMVLTVGIARELFPYQA-LTNLRNDAIATFLWVANWRFVV
MAB_2689|M.abscessus_ATCC_1997 LVRRLLPALILVLAACAVLTVLVQPETRWETFAEQSLASLGYYQNWELAN
Mflv_1859|M.gilvum_PYR-GCK QDTDYFSQGAPPSPLQHTWSLAVEEQYYVLWPLLIAAVIAGLTALAYRRG
Mvan_4875|M.vanbaalenii_PYR-1 QHTDYFSQGAPPSPLQHTWSLAVEEQFYVLWPLLIAAVLAGLAALAYRRG
MSMEG_5537|M.smegmatis_MC2_155 QQADYFSQGAPPSPLQHTWSLGVEEQYYLVWPLLVLFAVL----LLRRRG
TH_1216|M.thermoresistible__bu --------------------------------------------------
Mb0115|M.bovis_AF2122/97 QNTDYFTQGAPPSPLQHTWSLGVEEQYYVVWPLLLIGATLLLAARARRRC
Rv0111|M.tuberculosis_H37Rv QNTDYFTQGAPPSPLQHTWSLGVEEQYYVVWPLLLIGATLLLAARARRRC
MMAR_0312|M.marinum_M QKTDYFTQGAPPSPLQHTWSLGVEEQYYIVWPLLLIAVTLLLAARARRYF
MUL_4810|M.ulcerans_Agy99 QKTDYFTQGVPPSPLQHTWSLGVEEQYYIVWPLLLIAVTLLLAARARRYF
MAV_5201|M.avium_104 QKTDYFTQGAPPSPLQHTWSLGVEEQYYFVWPLLLIAVALALAARARRRC
MLBr_02670|M.leprae_Br4923 HKADYFTQGTPPSPLQHAWSLGVEEQYYLVWPLLLIAVTLLLAARARRRC
MAB_2689|M.abscessus_ATCC_1997 TAADYLAASESVSPLQHLWSMSVQGQFYVGFLALVYLLAVLLRKPLGRHI
Mflv_1859|M.gilvum_PYR-GCK MTLSQRAVRTAVFLLATAGIVASATATALMSSDGNLNRVYFGTDTRVQAL
Mvan_4875|M.vanbaalenii_PYR-1 VPLSQRVVRTAVFVLAAGGVVASATATFFLSSDATLNRVYFGTDTRVQAL
MSMEG_5537|M.smegmatis_MC2_155 A----RAVRIAVVVIATAGAAASAACAIVAVNTDTLNRVYFGTDFRAQGL
TH_1216|M.thermoresistible__bu --------------------------------------------------
Mb0115|M.bovis_AF2122/97 RRATVGGVRFAAFLIASLGTMASATAAVAFTSAATRDRIYFGTDTRAQAL
Rv0111|M.tuberculosis_H37Rv RRATVGGVRFAAFLIASLGTMASATAAVAFTSAATRDRIYFGTDTRAQAL
MMAR_0312|M.marinum_M QRATLGGVRFTIFVIGTLGALASGAAAIVFASESTRDRIYFGTDTRAQAL
MUL_4810|M.ulcerans_Agy99 QRATLGGVRFTIFVIGTLGALASGAAAIVFASESTRDRIYFGTDTRAQAL
MAV_5201|M.avium_104 TRATVGGVRFSAFVIATVGALASAAVAIVMAADGTRDRIYFGTDTRAQAL
MLBr_02670|M.leprae_Br4923 RYATLGAVRLAVFVLASLGALASATVAIARVCDVTRDRIYFGTDTRAQAL
MAB_2689|M.abscessus_ATCC_1997 R---------TVLIVVIAVLSAASFGYAIYAHLDFQSIAYYNTFARAWEL
Mflv_1859|M.gilvum_PYR-GCK LVGAAAAALLVRDWRGLTAGLEIFRSRWARWVAWSLPVVGLVVLFLAARI
Mvan_4875|M.vanbaalenii_PYR-1 LVGAAAAALVVRDWRGLTAGLVIFRSRWARWVAWSLPAVGLVVLFSAARM
MSMEG_5537|M.smegmatis_MC2_155 LLGAAAAALLVRDWSALTVSGTLIRARWRRWIAWVLPVFGLAVLGAAAHL
TH_1216|M.thermoresistible__bu --------------------------------------------------
Mb0115|M.bovis_AF2122/97 LIGSAAAALLVRDWPSLNRGWCLIRTRWGRRIARLLPFVGLAGLAVTTHV
Rv0111|M.tuberculosis_H37Rv LIGSAAAALLVRDWPSLNRGWCLIRTRWGRRIARLLPFVGLAGLAVTTHV
MMAR_0312|M.marinum_M LVGAAAAALLVRDWTALNHGWCLVRTRWGRRVARFLPVIGVAGLAALTHF
MUL_4810|M.ulcerans_Agy99 LVGAAAAALLVRDWTALNHGWCLVRTRWGRRVARFLPVIGVAGLAALTHF
MAV_5201|M.avium_104 LVGAAAAALLVSDWPSMNRGWCMLRTRWARRVARILPVLGLAGLAAATHC
MLBr_02670|M.leprae_Br4923 LVGAAASALLVRDWPSLNRGWCLIRTRWGQLIARFLPVLGLAGLAAATHY
MAB_2689|M.abscessus_ATCC_1997 LLGVLVGALVAG-------------TRWPRWLRQLLSFVAVVAILSCGAL
Mflv_1859|M.gilvum_PYR-GCK ATGSVEEFRSGLLIVVALAAVLVIAPVALA----QDGPVARLLSVWPLVG
Mvan_4875|M.vanbaalenii_PYR-1 ATGSAAEFRSGLLIVVALAAVLVIAPVALA----QDGPVARLLSAGPLVG
MSMEG_5537|M.smegmatis_MC2_155 ATGDAEDFRHGLLIIVAAAAVLVIVPVALD----QYGWVARALGWRPLVA
TH_1216|M.thermoresistible__bu --------------------------------------------------
Mb0115|M.bovis_AF2122/97 ATGSVGEFRHGLLIVVAGAAVIVVASVAME----QRGAVARILAWRPLVW
Rv0111|M.tuberculosis_H37Rv ATGSVGEFRHGLLIVVAGAAVIVVASVAME----QRGAVARILAWRPLVW
MMAR_0312|M.marinum_M ATGNVNEFRHGLLILVALAAVFVVAPVALE----QRGAVARLLAWRPLVW
MUL_4810|M.ulcerans_Agy99 ATGNVNEFRHGLLILVALAAVFVVAPVALE----QRGAVARLLAWRPLVW
MAV_5201|M.avium_104 ASGASGEFRHGLLIGVAIAAVAVIAPVAVE----QRGVVARVLALPPLVW
MLBr_02670|M.leprae_Br4923 ATGGAGDFRHGLLIGVAIAAVIVIAPVALE----QRGTVARLLALRPLVW
MAB_2689|M.abscessus_ATCC_1997 ING-VREFPGPLALVPVVATLLLILSAANLPADARQPVANRFLATRPLVE
Mflv_1859|M.gilvum_PYR-GCK LGAISYGVYLWHWPIFLILNGER-TGASGWQLFALRCAATIAVAAVSWWL
Mvan_4875|M.vanbaalenii_PYR-1 LGAISYGVYLWHWPIFLVLNGER-TGIEGWELFALRCAATVAVAAVSWWL
MSMEG_5537|M.smegmatis_MC2_155 LGAISYGVYLWHWPLFLILNGER-TGLTGWSLVALRFGVTIALAWVSWRF
TH_1216|M.thermoresistible__bu --------------------------------------------------
Mb0115|M.bovis_AF2122/97 LGTISYGVYLWHWPIFLALNGQR-TGWSGPALFAARCAATVVLAGASWWL
Rv0111|M.tuberculosis_H37Rv LGTISYGVYLWHWPIFLALNGQR-TGWSGPALFAARCAATVVLAGASWWL
MMAR_0312|M.marinum_M LGTISYGIYLWHWPIFLALNGER-TGWTGMRLFAARCALTVAIAALSWWL
MUL_4810|M.ulcerans_Agy99 LGTISYGIYLWHLPIFLALNGER-TGWTGMRLFAARCALTVAIAALSWWL
MAV_5201|M.avium_104 LGTISYGVYLWHWPIFLALNGER-TGWAGMPLFAARCAATVAVAAASWWL
MLBr_02670|M.leprae_Br4923 LGTISYGIYLWHWPIFLALNGER-TGWSGVPLFASRCSATVAVAAVSWWL
MAB_2689|M.abscessus_ATCC_1997 LGALAYSLYLWHWPLLVYWLVYTGEARVTVAEGAAILGISLALAYLTNKY
Mflv_1859|M.gilvum_PYR-GCK IEQPVRE----------WRPAFVPMLPLAGATAATAAVLTMT------VL
Mvan_4875|M.vanbaalenii_PYR-1 IEQPIRE----------WRPAFVPMLPLAAATAATAAVVTMT------VL
MSMEG_5537|M.smegmatis_MC2_155 VEQPVRR----------WRPQHVPMLPLAAATAATAAVVTLT------VV
TH_1216|M.thermoresistible__bu --------------------------------------------------
Mb0115|M.bovis_AF2122/97 IEQPIRR----------WRPARVPLLPLAAATVASAAAVTML------VV
Rv0111|M.tuberculosis_H37Rv IEQPIRR----------WRPARVPLLPLAAATVASAAAVTML------VV
MMAR_0312|M.marinum_M FEQPIRR----------WRPARVPLLPLAAATVASAAAATML------VI
MUL_4810|M.ulcerans_Agy99 FEQPIRR----------WRPARVPLLPLAAATVASAAAATML------VI
MAV_5201|M.avium_104 LEQPIRR----------WRPARVPLLPLAAATVASAAAVTLL------VV
MLBr_02670|M.leprae_Br4923 IEQPIRH----------WRPVRVPLLPLAAATVASAAAATMW------VI
MAB_2689|M.abscessus_ATCC_1997 IETPLRYPRVSPTSASLWTRLRRPTLALGTSIVLMAVALTATSFTWLEHV
Mflv_1859|M.gilvum_PYR-GCK PVRVVP--PTPEPGPLVDIAALVGPEVPVE---VTRAPTPRAPGDLRVAV
Mvan_4875|M.vanbaalenii_PYR-1 PVRMVP--ATPEPGPEVDIAALVGAEVPVE---VTRAPRPSAPGDLRVAV
MSMEG_5537|M.smegmatis_MC2_155 PVTPPQ--PTP-LGPDVMSAAAVQPDVGAERPVLGPAPARRAPGTHSVAV
TH_1216|M.thermoresistible__bu --------------------------------------------------
Mb0115|M.bovis_AF2122/97 PVGAGPGLREIGLPPGVSAVAAVSPSPPEASQPAPG--PRDPNRPFTVSV
Rv0111|M.tuberculosis_H37Rv PVGAGPGLREIGLPPGVSAVAAVSPSPPEASQPAPG--PRDPNRPFTVSV
MMAR_0312|M.marinum_M PVGTGPGLREVGLPPGVSAVAAVSPSPPEPAAPGPAAAPRDPNRPFTVSV
MUL_4810|M.ulcerans_Agy99 PVGNGPGLREVGLPPGVSAVAAVSPSPPEPAAPGPAAAPRDPNRPFTVSV
MAV_5201|M.avium_104 PVGAGPGLREAGLPPGVSAVAAVSPSPPGNSRP------RDPNRPFTVSV
MLBr_02670|M.leprae_Br4923 PVGTETGLRDIALPPEVSTVAAVSPSPPRGKLSS----TRDPNQPFTVAV
MAB_2689|M.abscessus_ATCC_1997 TVQRSNGKELSGLRPRDYPGAGALLYGDRVPDLPMRPTTLEASDDLPITT
Mflv_1859|M.gilvum_PYR-GCK FGDSIA--------------------------------WTMMRYLPDTPG
Mvan_4875|M.vanbaalenii_PYR-1 FGDSIA--------------------------------WTLMRYLPETPG
MSMEG_5537|M.smegmatis_MC2_155 FGDSVA--------------------------------WTLMRYLPATPG
TH_1216|M.thermoresistible__bu -----------------------------------------MRYLPPMPG
Mb0115|M.bovis_AF2122/97 FGDSIG--------------------------------WTLMHYLPPTPG
Rv0111|M.tuberculosis_H37Rv FGDSIG--------------------------------WTLMHYLPPTPG
MMAR_0312|M.marinum_M FGDSIG--------------------------------WTMMHYLPATPG
MUL_4810|M.ulcerans_Agy99 FGDSIG--------------------------------WTMMHYLPATPG
MAV_5201|M.avium_104 FGDSIG--------------------------------WTWMHYLPPTPG
MLBr_02670|M.leprae_Br4923 FGDSIG--------------------------------WTWMHYLPPTPG
MAB_2689|M.abscessus_ATCC_1997 EQDCISDFRNRAVITCTYGNPHATRTIALAGGSHAEHWITALDILGRQHN
: * .
Mflv_1859|M.gilvum_PYR-GCK VDFADFTTIGCGIARGGPYEYVGQELQQKPECDEWPARWAQRIGHQKPDV
Mvan_4875|M.vanbaalenii_PYR-1 LDFADYTTIGCGIARGGPYEYVGQELPQKPECDEWPARWAQRVGHEKPDV
MSMEG_5537|M.smegmatis_MC2_155 LAFSNYTTIGCGIARGGPYRSAGETLNQKPECDTWPARWTQRIRHDRPDT
TH_1216|M.thermoresistible__bu MTFIDRTTIGCGLARGGPYRQLGETLNQKPECDVWPARWTQRINYDKPDV
Mb0115|M.bovis_AF2122/97 FRFIDHTVIGCSLVRGTPYRYIGQTLEQRAECDGWPARWSAQVNRDQPDV
Rv0111|M.tuberculosis_H37Rv FRFIDHTVIGCSLVRGTPYRYIGQTLEQRAECDGWPARWSAQVNRDQPDV
MMAR_0312|M.marinum_M FRFIDHTVIGCSLVRGTPYRYIGQTLEQRPECDTWPSRWSAQVSQDRPDV
MUL_4810|M.ulcerans_Agy99 FRFIDHTVIGCSLVRGTPYRYIGQTLEQRPECDTWPSRWSAQVSQDRPDV
MAV_5201|M.avium_104 FAFLDHTVIGCSLVRGTPYRYIGETLEQRPECDGWPIRWSRQVSQDQPDV
MLBr_02670|M.leprae_Br4923 FAFIDHTVIGCSLVRGTPYQYIGQTLEQRPECDNWPIRWSTQINKDHPNV
MAB_2689|M.abscessus_ATCC_1997 FKITTYLKMGCPLSTEEVPRIAGSNDP-YPDCKKWVDEVMSRIIQEHPDY
. : :** : . *. .:*. * . :: ::*:
Mflv_1859|M.gilvum_PYR-GCK VLLMVGR-------WEVVDRMSEGRWTHIGEPGYDAYLRSELNRALDILG
Mvan_4875|M.vanbaalenii_PYR-1 VLLMVGR-------WEVVDRMMEGQWTHIGEAGYDAYLRGELNRALDILG
MSMEG_5537|M.smegmatis_MC2_155 VLLMIGR-------WEIVDRKHEGHWTHIGEDAYDDYLTAELQRALDILG
TH_1216|M.thermoresistible__bu ALLIVGR-------WEIVDRVFEGRWTHIGDPAYDAYLTGELRRALDIIS
Mb0115|M.bovis_AF2122/97 ALLIVGR-------WETVDRVNEGRWTHIGDPTFDAYLNAELQRALSIVG
Rv0111|M.tuberculosis_H37Rv ALLIVGR-------WETVDRVNEGRWTHIGDPTFDAYLNAELQRALSIVG
MMAR_0312|M.marinum_M ALLIIGR-------WETVDRVNEGKWTHIGDPTFDGYLNFELQRALKIVG
MUL_4810|M.ulcerans_Agy99 ALLIIGR-------WETVDRVNEGKWTHIGDPTFDGYLNFELQRALKIVG
MAV_5201|M.avium_104 ALLIIGR-------WETVDRVNEGQWTHIGDPTFDAYLNGELERALNIVG
MLBr_02670|M.leprae_Br4923 ALLIIGR-------WETVNRVNEGQWTHIGDPTFDSYLNAELQQALNILG
MAB_2689|M.abscessus_ATCC_1997 VFTTTTRPRSALGDGDVMPESYLGIWSAFDEAGIPVLGMRDTPWLIDKDG
.: * : : . * *: :.: : :. .
Mflv_1859|M.gilvum_PYR-GCK STGARLVVATEPYNRRAEKADGSLYPEDQPDRVERWNTMLREVVDERPNA
Mvan_4875|M.vanbaalenii_PYR-1 STGARVVVTTEPYNRRAEKPDGSLYPEDQPKRVETWNALLRSVVDKRPNA
MSMEG_5537|M.smegmatis_MC2_155 STGARVVVTTAPYTRRGERADGSLYPEDQPSRVRAWNTILRDVAAKRPNV
TH_1216|M.thermoresistible__bu STGARVIVTTVPYNRRGEKPDGSLYPEDQPERVDRWNRILRSVVDEYPGV
Mb0115|M.bovis_AF2122/97 STGVRVMVTTVPYSRGGEKPDGRLYPEDQPERVNKWNAMLHNAISQHSNV
Rv0111|M.tuberculosis_H37Rv STGVRVMVTTVPYSRGGEKPDGRLYPEDQPERVNKWNAMLHNAISQHSNV
MMAR_0312|M.marinum_M SSGVRVMVATVPYSRGGEKPDGRLYPEDQPDRVRQWNDMLRKTVSQHRNV
MUL_4810|M.ulcerans_Agy99 SSGVRVMVATVPYSRGGEKPDGRLYPEDQPDRVRQWNDMLRKTVSQHRNV
MAV_5201|M.avium_104 STGVRVVVATVPYSRGGEKPDGRLYPEDQPDRVNLWNTMLRKTVAHHPNV
MLBr_02670|M.leprae_Br4923 SSGVRVVVATVPYSRGGEKPDGQLYPEDQPDRVNQWNTMLRNTVSQHSNV
MAB_2689|M.abscessus_ATCC_1997 TT-----YTAADCISAGGTADS--CAMDRNRALDDVNPTLAIAN-RFPLL
:: :: . .*. . *: : * * . .
Mflv_1859|M.gilvum_PYR-GCK TVLDLNEKLSPNGYYQMRVNGLKVRSDGVHPTPEAVEWMTPWLVEAIRTP
Mvan_4875|M.vanbaalenii_PYR-1 TVLDLNKKLSPNGYYQTRVDGIRVRSDGVHPTPEAVEWLTPWLVDALKPE
MSMEG_5537|M.smegmatis_MC2_155 SVLDFNAKLNPNGAYTARVDGIRMRSDGVHPTAEAVEWLTPWLVDSLKKL
TH_1216|M.thermoresistible__bu SVLDLNKKLGPNGRYTAKVDGVRVRSDGVHFTPEGVEWLAPWLQEAVAPP
Mb0115|M.bovis_AF2122/97 GMIDLNKKLCPDGVYTAKVDGIKVRSDGVHLTQEGVKWLIPWLEDSVRVA
Rv0111|M.tuberculosis_H37Rv GMIDLNKKLCPDGVYTAKVDGIKVRSDGVHLTQEGVKWLIPWLEDSVRVA
MMAR_0312|M.marinum_M GIIDLNKKLSPDGVYTAKVDGIQVRSDGVHLTPEGVKWLIPWLEESLR--
MUL_4810|M.ulcerans_Agy99 GIIDLNKKLSPDGVYTAKVDGIQVRSDGVHLTPEGVKWLIPWLEESLR--
MAV_5201|M.avium_104 AILDLNKKLCPDGVYTAKVDGIKVRSDGVHLTPEGVKWLTPWLEESLR--
MLBr_02670|M.leprae_Br4923 QILDLNKKLCPDGVYTAKVDGIKVRSDGVHLTPEGVKWLTPWLEESLK--
MAB_2689|M.abscessus_ATCC_1997 KILDMTRAICRPDKCRVVEGNVLVYHDSHHISATYMRTMAKELGRQIALA
::*:. : . ..: : *. * : :. : * :
Mflv_1859|M.gilvum_PYR-GCK ------------------------------
Mvan_4875|M.vanbaalenii_PYR-1 ------------------------------
MSMEG_5537|M.smegmatis_MC2_155 PPGKPR------------------------
TH_1216|M.thermoresistible__bu NTPAPDKPAPEPATPKPPHKPPHNNRPVAE
Mb0115|M.bovis_AF2122/97 S-----------------------------
Rv0111|M.tuberculosis_H37Rv S-----------------------------
MMAR_0312|M.marinum_M ------------------------------
MUL_4810|M.ulcerans_Agy99 ------------------------------
MAV_5201|M.avium_104 ------------------------------
MLBr_02670|M.leprae_Br4923 ------------------------------
MAB_2689|M.abscessus_ATCC_1997 TGWWRPAQPGQ-------------------