For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGWVAKIFRVGRVVEPAAPLPAAIAEPPAGVRGSLQIRHVDAGSCNGCEVEISGAFGPVYDAERFGARLV ASPRHADALLVTGVVTHNMAGPLRKTLEATPRPRVVIACGDCALNRGVFADAYGVVGAVGEVVPVDVEIA GCPPTPAAIMAALRSVTGK
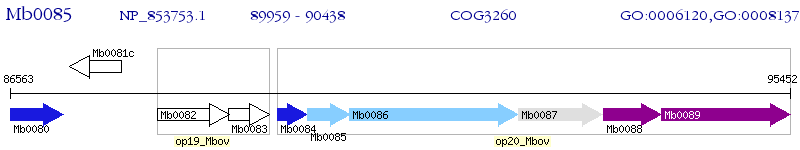
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0085 | - | - | 100% (159) | oxidoreductase |
| M. bovis AF2122 / 97 | Mb3170 | nuoB | 1e-17 | 40.19% (107) | NADH dehydrogenase subunit B |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4482 | - | 6e-18 | 39.82% (113) | NADH dehydrogenase subunit B |
| M. tuberculosis H37Rv | Rv0082 | - | 4e-89 | 99.37% (159) | oxidoreductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2135 | - | 1e-17 | 40.19% (107) | NADH dehydrogenase subunit B |
| M. marinum M | MMAR_1654 | - | 4e-71 | 79.25% (159) | oxidoreductase |
| M. avium 104 | MAV_5114 | - | 1e-67 | 89.55% (134) | hydrogenase-4, I subunit, putative |
| M. smegmatis MC2 155 | MSMEG_2062 | nuoB | 1e-17 | 39.82% (113) | NADH dehydrogenase subunit B |
| M. thermoresistible (build 8) | TH_0636 | nuoB | 4e-17 | 38.05% (113) | PROBABLE NADH DEHYDROGENASE I (CHAIN B) NUOB |
| M. ulcerans Agy99 | MUL_1891 | - | 1e-69 | 77.99% (159) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_1884 | - | 6e-18 | 39.82% (113) | NADH dehydrogenase subunit B |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4482|M.gilvum_PYR-GCK ---------MGLEEKLPGGILLSTVEKVAGYVRKGSLWPATFG-LACCAI
Mvan_1884|M.vanbaalenii_PYR-1 ---------MGLEERLPGGILLSTVEKVAGYVRKGSLWPATFG-LACCAI
MSMEG_2062|M.smegmatis_MC2_155 ---------MGLEERLPGGILLSTVETVAGYVRKGSLWPATFG-LACCAI
TH_0636|M.thermoresistible__bu ---------VGLEEQLPGGILLSTVEKVAGYVRKGSLWPATFG-LACCAI
MAB_2135|M.abscessus_ATCC_1997 ---------MGLEEKLPSGFLLSTVETLAGYVRKGSLWPATFG-LACCAI
Mb0085|M.bovis_AF2122/97 MGWVAKIFRVGRVVEPAAPLPAAIAEPPAGVRGSLQIRHVDAGSCNGCEV
Rv0082|M.tuberculosis_H37Rv MGWVAKIFRVGRVVEPAAPLPAAIAEPPAGVRGSLQIRHVDAGSCNGCEV
MAV_5114|M.avium_104 ------------------------MEPPAGVEGSLQVRHVDAGSCNGCEV
MMAR_1654|M.marinum_M MGWIARIMRLGRVAEKLGDESTPAVAAPAGLRGSLQVRHVDAGSCNGCEV
MUL_1891|M.ulcerans_Agy99 MGWIARIMRLGLVAEKLGDESTPAVAAPAGLRGSLQVRHVDAGSCNGCEV
** . .: . * * :
Mflv_4482|M.gilvum_PYR-GCK EMMATAGPRFDISRFGMERFSATPRQADLMIVAGRVSQKMAPVLRQIYDQ
Mvan_1884|M.vanbaalenii_PYR-1 EMMATAGPRFDISRFGMERFSATPRQADLMIVAGRVSQKMAPVLRQIYDQ
MSMEG_2062|M.smegmatis_MC2_155 EMMSTAGPRFDIARFGMERFSATPRQADLMIVAGRVSQKMAPVLRQIYDQ
TH_0636|M.thermoresistible__bu EMMATASPRFDIARFGMERFSATPRQADLMIVAGRVSQKMAPVLRQIYDQ
MAB_2135|M.abscessus_ATCC_1997 EMMSTAGPRFDIARFGMERFSATPRQADLMIVAGRVSQKMAPVLRQIYDQ
Mb0085|M.bovis_AF2122/97 EISGAFGPVYDAERFG-ARLVASPRHADALLVTGVVTHNMAGPLRKTLEA
Rv0082|M.tuberculosis_H37Rv EISGAFGPVYDAERFG-ARLVASPQHADALLVTGVVTHNMAGPLRKTLEA
MAV_5114|M.avium_104 EISGAFGPVYDAERFG-ARLVASPRHADALLVTGVVTRNMVEPLRNTLAA
MMAR_1654|M.marinum_M EISGAFGPVYDAERFG-ARLVASPRHADALLVTGVVTRNMAQPLKNTLAA
MUL_1891|M.ulcerans_Agy99 EISGAFGPVYDAERVG-ARLVASPRHADALLVTGVVTRNMAQPLKNTLAA
*: .: .* :* *.* *: *:*::** ::*:* *:::*. *::
Mflv_4482|M.gilvum_PYR-GCK MAEPKWVLAMGVCASSGGMFNN-YAVVQGVDHVVPVDIYLPGCPPRPEML
Mvan_1884|M.vanbaalenii_PYR-1 MAEPKWVLAMGVCASSGGMFNN-YAVVQGVDHVVPVDIYLPGCPPRPEML
MSMEG_2062|M.smegmatis_MC2_155 MVEPKWVLAMGVCASSGGMFNN-YAVVQGVDHVVPVDIYLPGCPPRPEML
TH_0636|M.thermoresistible__bu MAEPKWVLAMGVCASSGGMFNN-YAIVQGVDHVVPVDIYLPGCPPRPEML
MAB_2135|M.abscessus_ATCC_1997 MAEPKWVLAMGVCASSGGMFNN-YAIVQGVDHVVPVDIYLPGCPPRPEML
Mb0085|M.bovis_AF2122/97 TPRPRVVIACGDCALNRGVFADAYGVVGAVGEVVPVDVEIAGCPPTPAAI
Rv0082|M.tuberculosis_H37Rv TPRPRVVIACGDCALNRGVFADAYGVVGAVGEVVPVDVEIAGCPPTPAAI
MAV_5114|M.avium_104 TPQPRVVIACGDCALNRGVFTDAYGVVGAVGEIVPVDVEVAGCPPTPAQI
MMAR_1654|M.marinum_M TPQPRVVIACGDCALNRGVFADAYGVVGAVSEVVPVDVEVPGCPPTPDQV
MUL_1891|M.ulcerans_Agy99 TPQPRVVIACGDCALNRGVFADAYGVVGAVSEVVPVDVEVPGCPPTPDQV
.*: *:* * ** . *:* : *.:* .*..:****: :.**** * :
Mflv_4482|M.gilvum_PYR-GCK LHAILKLHDKIQQMPLGVNREEAIREAEQAAMALTPTIELKGLLR
Mvan_1884|M.vanbaalenii_PYR-1 LHAILKLHDKIQQMPLGVNREEAIREAERAALAVTPTIELKGLLR
MSMEG_2062|M.smegmatis_MC2_155 LHAILKLHDKIQQMPLGVNREEAIREAEQAALAVPPTIELKGLLR
TH_0636|M.thermoresistible__bu LHAILKLHDKIQQMPLGVHRDEVIRETERAALAAPTTVEFKGRMR
MAB_2135|M.abscessus_ATCC_1997 LHAILKLHEKIAHMPLGANREEVIRETEAAALAATPTIEMKGLLR
Mb0085|M.bovis_AF2122/97 MAALRSVTGK-----------------------------------
Rv0082|M.tuberculosis_H37Rv MAALRSVTGK-----------------------------------
MAV_5114|M.avium_104 VDALRSVTGK-----------------------------------
MMAR_1654|M.marinum_M VAALRSVTGR-----------------------------------
MUL_1891|M.ulcerans_Agy99 VAALRSVTGR-----------------------------------
: *: .: :