For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TITYDEALRLRIRARLTSHERRAVTDPTKRHAAVAVVLVDSEVGEDRVDPAPVDEWIGGRLMPEPGLDGR MVDVSGGAAFFLCRRASRLRAHAAQWALPGGRVDPGENAIEAALRELDEEIGIRLPDSAVLGLLDDYPTR SGYVITPVVIWGGGRLDPHLVPDEVVAVYRVGLHQLQRQDSPRFVAIPESPRPVVQVPLGNDLIHAPTGA VLLQVRWLCLEGRDDPVDELEQPVFAWK*
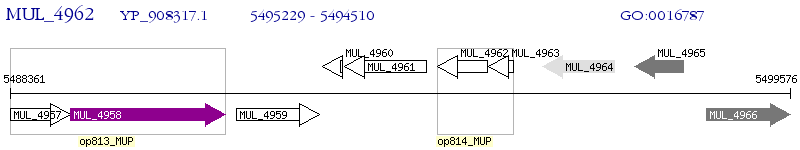
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4962 | - | - | 100% (239) | hypothetical protein MUL_4962 |
| M. ulcerans Agy99 | MUL_1008 | mutT2 | 3e-05 | 37.35% (83) | mutator protein MutT2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3696c | - | 6e-06 | 49.21% (63) | hypothetical protein Mb3696c |
| M. gilvum PYR-GCK | Mflv_1043 | - | 1e-114 | 82.01% (239) | NUDIX hydrolase |
| M. tuberculosis H37Rv | Rv3672c | - | 6e-06 | 49.21% (63) | hypothetical protein Rv3672c |
| M. leprae Br4923 | MLBr_02299 | - | 1e-05 | 39.47% (76) | hypothetical protein MLBr_02299 |
| M. abscessus ATCC 19977 | MAB_4210 | - | 1e-112 | 80.17% (237) | putative MutT/NUDIX family protein |
| M. marinum M | MMAR_0135 | - | 1e-136 | 98.74% (239) | hypothetical protein MMAR_0135 |
| M. avium 104 | MAV_1301 | - | 2e-06 | 46.30% (54) | hydrolase, NUDIX family protein |
| M. smegmatis MC2 155 | MSMEG_6617 | - | 1e-114 | 82.43% (239) | nudix hydrolase |
| M. thermoresistible (build 8) | TH_1236 | - | 1e-107 | 78.24% (239) | nudix hydrolase |
| M. vanbaalenii PYR-1 | Mvan_5787 | - | 1e-113 | 81.17% (239) | NUDIX hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4962|M.ulcerans_Agy99 ------------------MTITYDEALRLRIRARLTSHERRAVTDPTKRH
MMAR_0135|M.marinum_M ------------------MTITYDEALRLRIRARLTSHERRAVTDPTKRH
MAB_4210|M.abscessus_ATCC_1997 -------------------MIAYDEELRNRIRAHLARHERRTVDDPAKRR
MSMEG_6617|M.smegmatis_MC2_155 ------------------MTIAYDDALRERIRTHLAGHDRRVTTDPTKRH
Mflv_1043|M.gilvum_PYR-GCK ------------------MTLGYDDVLRDRIAQRLGQHTRRTVTDPSKRH
Mvan_5787|M.vanbaalenii_PYR-1 ------------------MTITYDDRLRDQLQQRLAGHARRTVSDPSKRH
TH_1236|M.thermoresistible__bu --------------MVEQVTIPYDAELRESIRRNLAGHDRRVVTDPTKRH
Mb3696c|M.bovis_AF2122/97 MSAGGTPLQAGATPTGSRGTVALRPDAGPSWLRPLVDNVGQIPDAYRRRL
Rv3672c|M.tuberculosis_H37Rv MSAGGTPLQAGATPTGSRGTVALRPDAGPSWLRPLVDNVGQIPDAYRRRL
MLBr_02299|M.leprae_Br4923 -----MPVSAGCTAR-VQGTVPLVFDVGPSWLRPLVDNIAVMPGVYQRLL
MAV_1301|M.avium_104 ---------------------------------------MRADGIPSLAM
MUL_4962|M.ulcerans_Agy99 AAVAVVLVDSEVGEDRVDPAPVDEWIGGRLMPEPGLDGRMVD--VSGGAA
MMAR_0135|M.marinum_M AAVAVVLVDSEVGEDRVDPAPVDEWIGGRPMPEPGLDGRMVD--VSGGAA
MAB_4210|M.abscessus_ATCC_1997 AAVAVVLVDSDLGEDRIDPAPLEEWIGGREIPGHGLDGRMVD--VSGGAA
MSMEG_6617|M.smegmatis_MC2_155 AAVAVVLVDSELGEDRVDPAPVEDWIGGRPVLEAGLDGRMVD--VSGGAA
Mflv_1043|M.gilvum_PYR-GCK AAVAVVLVDSVVGEDRVDPAPVDDWIAGRPMPD-DLDGRMVD--VSGGAA
Mvan_5787|M.vanbaalenii_PYR-1 AAVAVVLVDSLAGEDRVDPAPVDDWIAGRPMPE-DLDGRMVD--VSGGAA
TH_1236|M.thermoresistible__bu AAVAIVLVDSEVGEDRVDPAPVDDWNAGRGLSAPDLDGRMVD--VSGGAA
Mb3696c|M.bovis_AF2122/97 PADVLAMVTAAGAVSAMTSSRRDHREAAVLVLFSGPEAGPGDGGVPDDAD
Rv3672c|M.tuberculosis_H37Rv PADVLAMVTAAGAVSAMTSSRRDHREAAVLVLFSGPEAGPGDGGVPDDAD
MLBr_02299|M.leprae_Br4923 PADVLAIVTAARAIPSVA-LRRDGREAAVLVLFSGPESGPPDGGAPDDAD
MAV_1301|M.avium_104 PTQIVVAGALIRG------------------------------------S
.: :. .
MUL_4962|M.ulcerans_Agy99 FFLCRRASRLRAHAAQWALPGGRVDPGEN-AIEAALRELDEEIGIRLPDS
MMAR_0135|M.marinum_M FFLCRRASRLRAHAAQWALPGGRVDPGET-AIEAALRELDEEIGIRLPDS
MAB_4210|M.abscessus_ATCC_1997 FLLCRRASRLNSHAAQWALPGGRLDPGED-AIQAALRELDEELGVSLPDS
MSMEG_6617|M.smegmatis_MC2_155 FLMCRRATRLTSHSAQWALPGGRVDPGET-VEEAALRELDEELGVRLPES
Mflv_1043|M.gilvum_PYR-GCK FLLCRRASRLSSHSAQWALPGGRLDPGET-VVEAALRELDEEVGVRLPES
Mvan_5787|M.vanbaalenii_PYR-1 FFLCRRASRLSSHSAQWALPGGRLDPGET-VVDAALRELDEEVGVRLTET
TH_1236|M.thermoresistible__bu FVLCRRAFRLSSHSAQWALPGGRVDPGET-IVEAALRETHEEVGVTLPES
Mb3696c|M.bovis_AF2122/97 LLLTVRASTLRHHAGQAAFPGGVVDPADDGPVATALREANEETGIDPSRL
Rv3672c|M.tuberculosis_H37Rv LLLTVRASTLRHHAGQAAFPGGVVDPADDGPVATALREANEETGIDPSRL
MLBr_02299|M.leprae_Br4923 LLLTVRASTLRHHAGQAAFPGGASDPTDDGPVATALREAHEETGIDLARL
MAV_1301|M.avium_104 RLLVAQRARPPELAGRWELPGGKVAPGET-ERDALARELAEELGLRVGDI
.: : :.: :*** * : : ** ** *:
MUL_4962|M.ulcerans_Agy99 AVLGLLD-DYPTRSGYVITPVVIWGGGRLDPHLV-PDEVVAVYRVGLHQL
MMAR_0135|M.marinum_M AVLGLLD-DYPTRSGYVITPVVIWGGGRLDPHPV-PDEVVAVYRVGLHQL
MAB_4210|M.abscessus_ATCC_1997 TVLGLLD-DYPTRSGYVITPVVVWGGGRLELSPS-PDEVVAAYRVGLHQL
MSMEG_6617|M.smegmatis_MC2_155 TVLGLLD-DYPTRSGYVITPVVIWGGGRLELSPS-PDEVVAAYRVGLHQL
Mflv_1043|M.gilvum_PYR-GCK ALLGLLD-DYSTRSGYVITPVVLWGGGRLDPRPA-PDEVVAVYRVGLHQL
Mvan_5787|M.vanbaalenii_PYR-1 SVLGLLD-DYPTRSGYVITPVVIWGGGRLDPRPA-PDEVVAVYRVGLHQL
TH_1236|M.thermoresistible__bu SVLGLLD-DYPTRSGYVITPVVIWGGGRLDLRPA-PDEVVAAYRVGLHQL
Mb3696c|M.bovis_AF2122/97 HPLATMERTFIAPSRFHVVPVLAYSPDPGPVAVVNEAETAIVARVPVRAF
Rv3672c|M.tuberculosis_H37Rv HPLATMERTFIAPSRFHVVPVLAYSPDPGPVAVVNEAETAIVARVPVRAF
MLBr_02299|M.leprae_Br4923 HPLATLERMFIAPSRFHVVPVLAYSADPGPVAVVNEAETAIVARVPLCAF
MAV_1301|M.avium_104 AVGDRLGGDIAVDGGMTLR----------------------AYRVRLLRG
: . . : . ** :
MUL_4962|M.ulcerans_Agy99 QRQDSPRFVAIPESPR--PVVQVPLGNDLIHAPTGAVLLQVRWLCLEGRD
MMAR_0135|M.marinum_M QRQDSPRFVAIPESPR--PVVQVPLGNDLIHAPTGAVLLQVRWLCLEGRD
MAB_4210|M.abscessus_ATCC_1997 QREDSPRYITIPESAR--PVVQIPLGNDLIHAPTGAVLLQVRWLCLEGSG
MSMEG_6617|M.smegmatis_MC2_155 QREDSPRFVAIPESPR--PVVQIPLGNDLIHAPTGAVLLQVRRLCLEGRP
Mflv_1043|M.gilvum_PYR-GCK LREDSPRFITIPESPR--PVVQVPLGNDLIHAPTGAVLLQLRWLCLEGRD
Mvan_5787|M.vanbaalenii_PYR-1 LREDSPRFITIPESPR--PVVQMPLGNDLIHAPTGAVLLQLRWLGLEGRS
TH_1236|M.thermoresistible__bu QRPDSPRFVSIPESDR--PVVQIPLGGDLIHAPTGAVLLQLRLLGLEGRP
Mb3696c|M.bovis_AF2122/97 INPANRLMVYRRPHTRRWAGPAFLLNQMLVWGFTGQVISAVLDVAGWAQP
Rv3672c|M.tuberculosis_H37Rv INPANRLMVYRRPHTRRWAGPAFLLNQMLVWGFTGQVISAVLDVAGWAQP
MLBr_02299|M.leprae_Br4923 INPANRLMVYRRAHGHRWYGPAFLLNDMLVWGFTGQVIAAMLDVAGWAQS
MAV_1301|M.avium_104 -RPDAR-------------------DHRALRWITAAQLHDLDWVPADRG-
. . : *. : : :
MUL_4962|M.ulcerans_Agy99 DPVDELEQPVFAWK---------
MMAR_0135|M.marinum_M DPVDELEQPVFAWK---------
MAB_4210|M.abscessus_ATCC_1997 DRVDDLEQPVFAWR---------
MSMEG_6617|M.smegmatis_MC2_155 DPVYDLEQPVFAWK---------
Mflv_1043|M.gilvum_PYR-GCK DPVNELEQPVFAWR---------
Mvan_5787|M.vanbaalenii_PYR-1 DPVDELEQPVFAWK---------
TH_1236|M.thermoresistible__bu DPVHDLEQPVFAWR---------
Mb3696c|M.bovis_AF2122/97 WDTGDIRELDAAMVLIDDESDPR
Rv3672c|M.tuberculosis_H37Rv WDTGDIRELDAAMVLIDDESDPR
MLBr_02299|M.leprae_Br4923 WDVTDVREVGEAMALVDSQGDPR
MAV_1301|M.avium_104 -WLGDLARVL-------------
:: .