For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STDHIRRPIRRALISVYDKTGLVDLAQGLTAAGVEIVSTGSTAKTIASKGIPVTRVEELTGFPEVLDGRV KTLHPRVHAGLLADLRNPEHEAALAELGVEAFELVVVNLYPFSQTVESGASVDECVEQIDIGGPSMVRAA AKNHPSVAVVTDPQGYDGVLAAVRSGGFTFAERKKLASLAFQHTAEYDIAVASWMQSTVAPEQPETDFPR WFGRNWRRQAMLRYGENPHQQAALYADPGAWPGLAQAEQLHGKEMSYNNFTDADAAWRAAFDHEQKCVAI IKHANPCGIAISEVSVADAHRKAHACDPLSAYGGVIACNTEVSPEMAEYVSTIFTEVIVAPAYAPAALDQ LTKKKNIRVLVASEPQDGGAELRPISGGLLMQQRDQLDAAGDNPANWILATGSPADPATLTDLVFAWRSC RAVKSNAIVIVADGATIGVGMGQVNRVDAARLAVERGGERVNGAVAASDAFFPFPDGLETLTAAGVKAIV HPGGSVRDEEVTAAAAKAGITLYLTGSRHFVH*
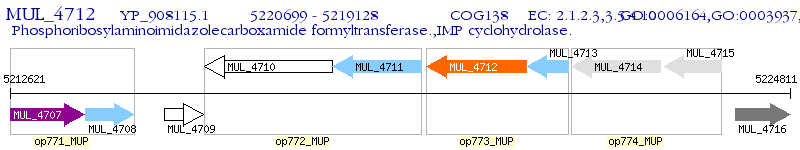
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4712 | purH | - | 100% (523) | bifunctional phosphoribosylaminoimidazolecarboxamide formyltransferase/IMP cyclohydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0982 | purH | 0.0 | 87.57% (523) | bifunctional phosphoribosylaminoimidazolecarboxamide |
| M. gilvum PYR-GCK | Mflv_1875 | purH | 0.0 | 78.72% (531) | bifunctional phosphoribosylaminoimidazolecarboxamide |
| M. tuberculosis H37Rv | Rv0957 | purH | 0.0 | 87.57% (523) | bifunctional phosphoribosylaminoimidazolecarboxamide |
| M. leprae Br4923 | MLBr_00161 | purH | 0.0 | 85.58% (520) | bifunctional phosphoribosylaminoimidazolecarboxamide |
| M. abscessus ATCC 19977 | MAB_1064 | - | 0.0 | 72.34% (517) | bifunctional phospho ribosylaminoimidazole carboxamide |
| M. marinum M | MMAR_4542 | purH | 0.0 | 99.24% (523) | bifunctional purine biosynthesis protein PurH |
| M. avium 104 | MAV_1081 | purH | 0.0 | 86.15% (527) | bifunctional phosphoribosylaminoimidazolecarboxamide |
| M. smegmatis MC2 155 | MSMEG_5515 | purH | 0.0 | 79.39% (524) | bifunctional phosphoribosylaminoimidazolecarboxamide |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4856 | purH | 0.0 | 80.46% (522) | bifunctional phosphoribosylaminoimidazolecarboxamide |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1875|M.gilvum_PYR-GCK ------MSDND------FRRPIRRALISVYDKSGLEALAQGLHAAGVDIV
Mvan_4856|M.vanbaalenii_PYR-1 ------MSDNDD----LFRRPIRRALISVYDKTGLVPLAQGLHAAGVDIV
MUL_4712|M.ulcerans_Agy99 ------MSTDH------IRRPIRRALISVYDKTGLVDLAQGLTAAGVEIV
MMAR_4542|M.marinum_M ------MSTDH------IRRPIRRALISVYDKTGLVDLAQGLTAAGVEIV
Mb0982|M.bovis_AF2122/97 ------MSTDD------GRRPIRRALISVYDKTGLVDLAQGLSAAGVEII
Rv0957|M.tuberculosis_H37Rv ------MSTDD------GRRPIRRALISVYDKTGLVDLAQGLSAAGVEII
MLBr_00161|M.leprae_Br4923 ------MSSDGER--DMARKPIRRALISVYDKTGLIDLAQGLNAAGVDIV
MAV_1081|M.avium_104 MKGDQPMSTDDWR--ENAKRPIRRALISVYDKTGLVELAQGLTEAGIEIV
MSMEG_5515|M.smegmatis_MC2_155 ------MSDKK---------PVRRALISVYDKTGLADLARGLHQAGVAIV
MAB_1064|M.abscessus_ATCC_1997 ------MERKKGLHQVSTQRPVRRALISVYDKTGLEELASGLHAAGVELV
*. . *:**********:** ** ** **: ::
Mflv_1875|M.gilvum_PYR-GCK STGSTAKNIAAAGIPVTPVEDVTGFPEVLDGRVKTLHPHVHAGLLADQRK
Mvan_4856|M.vanbaalenii_PYR-1 STGSTAKTIAGAGIPVTPVEDVTGFPEVLDGRVKTLHPHVHAGLLADQRK
MUL_4712|M.ulcerans_Agy99 STGSTAKTIASKGIPVTRVEELTGFPEVLDGRVKTLHPRVHAGLLADLRN
MMAR_4542|M.marinum_M STGSTAKTIASKGIPVTRVEELTGFPEVLDGRVKTLHPRVHAGLLADLRN
Mb0982|M.bovis_AF2122/97 STGSTAKTIADTGIPVTPVEQLTGFPEVLDGRVKTLHPRVHAGLLADLRK
Rv0957|M.tuberculosis_H37Rv STGSTAKTIADTGIPVTPVEQLTGFPEVLDGRVKTLHPRVHAGLLADLRK
MLBr_00161|M.leprae_Br4923 STGSTAKAIADQGIAVTPVEELTGFPEVLDGRVKTLHPRVHAGLLADLRK
MAV_1081|M.avium_104 STGSTAKVIAEKGIPVTRVEELTGFPEVLDGRVKTLHPRVHAGLLADLRK
MSMEG_5515|M.smegmatis_MC2_155 STGSTAKTIAAAGVPVTPVEQVTGFPEVLDGRVKTLHPRVHAGLLADTRK
MAB_1064|M.abscessus_ATCC_1997 STGSTAKTIAAASIPVTPVEDVTGFPEVLDGRVKTLHPHVHAGVLADTRR
******* ** .:.** **::****************:****:*** *.
Mflv_1875|M.gilvum_PYR-GCK AEHVTALEQLGVAAFELVVVNLYPFTQTVNSGAGVDECVEQIDIGGPSMV
Mvan_4856|M.vanbaalenii_PYR-1 AEHVAALAELGVTAFELVVVNLYPFTQTVNSGADEDECVEQIDIGGPSMV
MUL_4712|M.ulcerans_Agy99 PEHEAALAELGVEAFELVVVNLYPFSQTVESGASVDECVEQIDIGGPSMV
MMAR_4542|M.marinum_M PEHEAALAELGVEAFELVVVNLYPFSQTVESGASVDECVEQIDIGGPSMV
Mb0982|M.bovis_AF2122/97 SEHAAALEQLGIEAFELVVVNLYPFSQTVESGASVDDCVEQIDIGGPAMV
Rv0957|M.tuberculosis_H37Rv SEHAAALEQLGIEAFELVVVNLYPFSQTVESGASVDDCVEQIDIGGPAMV
MLBr_00161|M.leprae_Br4923 PEHAAALEQLGIEAFELVVVNLYPFSQTVKSGATVDECVEQIDIGGSSMV
MAV_1081|M.avium_104 PEHAAALEQLGIAAFELVVVNLYPFTETVDSGAGIDECVEQIDIGGPSMV
MSMEG_5515|M.smegmatis_MC2_155 PEHLAALKELDVEAFELVVVNLYPFSETVASGASVDECVEQIDIGGPSMV
MAB_1064|M.abscessus_ATCC_1997 EEHLTQLEELGVQGFELVVVNLYPFAATVASGASEDECVEQIDIGGPSMV
** : * :*.: .***********: ** *** *:*********.:**
Mflv_1875|M.gilvum_PYR-GCK RAAAKNHPSVAVVVDPLGYDGVLAAVRAGGFTYTERKKLAALAFRHTAEY
Mvan_4856|M.vanbaalenii_PYR-1 RAAAKNHPSVAVVVDPLGYDGVLAAVRAGGFTYSERKKLAALAFRHTAEY
MUL_4712|M.ulcerans_Agy99 RAAAKNHPSVAVVTDPQGYDGVLAAVRSGGFTFAERKKLASLAFQHTAEY
MMAR_4542|M.marinum_M RAAAKNHPSVAVVTDPQGYDGVLAAVSSGGFTLAERKKLASLAFQHTAEY
Mb0982|M.bovis_AF2122/97 RAAAKNHPSAAVVTDPLGYHGVLAALRAGGFTLAERKRLASLAFQHIAEY
Rv0957|M.tuberculosis_H37Rv RAAAKNHPSAAVVTDPLGYHGVLAALRAGGFTLAERKRLASLAFQHIAEY
MLBr_00161|M.leprae_Br4923 RAAAKNHPSVAVVTDPLGYVGVLAAVQGGGFTLAERKMLASMAFQHIAEY
MAV_1081|M.avium_104 RAAAKNHPSVAVVVDPLGYDGVLAAVRHGGFTLAERKRLAALAFQHTADY
MSMEG_5515|M.smegmatis_MC2_155 RAAAKNHPSVAVVVDPLGYDGVLAAVRAGGFTLAERKKLASVAFRHTAEY
MAB_1064|M.abscessus_ATCC_1997 RAAAKNHPSVAVVVDPAAYGEVLAAVTAGGFTLEARKVLAAKAFRHTAEY
*********.***.** .* ****: **** ** **: **:* *:*
Mflv_1875|M.gilvum_PYR-GCK DVAVASWMGSVLAPEDDAPEGAADDVAQLPPWLGSTFRRSAVLRYGENPH
Mvan_4856|M.vanbaalenii_PYR-1 DVAVASWMESVLAPEAEATSG------DLPPWLGATFRRAAVLRYGENPH
MUL_4712|M.ulcerans_Agy99 DIAVASWMQSTVAPEQ--------PETDFPRWFGRNWRRQAMLRYGENPH
MMAR_4542|M.marinum_M DIAVASWMQSTVAPEQ--------PETDFPRWFGRNWRRQAMLRYGENPH
Mb0982|M.bovis_AF2122/97 DIAVASWMQQTLAPEH--------PVAAFPQWFGRSWRRVAMLRYGENPH
Rv0957|M.tuberculosis_H37Rv DIAVASWMQQTLAPEH--------PVAAFPQWFGRSWRRVAMLRYGENPH
MLBr_00161|M.leprae_Br4923 EIAVASWMQSTLAPEQ--------PPTAFPQWFGRSWRRSAILRYGENPH
MAV_1081|M.avium_104 DIAVATWMESTLAPEH--------PPTTFPKWLGRSWRRSAMLRYGENPH
MSMEG_5515|M.smegmatis_MC2_155 DVAVASWMGSTLSPEEGSAEGGA-VEPSLPQWFAGTWHRAAVLRYGENPH
MAB_1064|M.abscessus_ATCC_1997 DVAVAGWLSGVAEPGD----------AELPSWIGGTWNRSAVLRYGENPH
::*** *: . * :* *:. .:.* *:********
Mflv_1875|M.gilvum_PYR-GCK QKAALYGDEGGWPGLAQAEQLHGKEMSYNNYTDADAAWRAAFDQEAICVA
Mvan_4856|M.vanbaalenii_PYR-1 QQAALYRDDGGWPGLAQAEQLHGKEMSYNNYTDADAAWRAAFDHEDICVA
MUL_4712|M.ulcerans_Agy99 QQAALYADPGAWPGLAQAEQLHGKEMSYNNFTDADAAWRAAFDHEQKCVA
MMAR_4542|M.marinum_M QQAALYADPGAWPGLAQAEQLHGKEMSYNNFTDADAAWRAAFDHEQKCVA
Mb0982|M.bovis_AF2122/97 QQAALYGDPTAWPGLAQAEQLHGKDMSYNNFTDADAAWRAAFDHEQTCVA
Rv0957|M.tuberculosis_H37Rv QQAALYGDPTAWPGLAQAEQLHGKDMSYNNFTDADAAWRAAFDHEQTCVA
MLBr_00161|M.leprae_Br4923 QQAALYSDPSACPGLAQAEQLHGKNMSYNNFTDADAAWRAAFDHEQSCVA
MAV_1081|M.avium_104 QQASLYSDPGAWPGLAQAEQLHGKEMSYNNFTDADAAWRAAFDHEQTCVA
MSMEG_5515|M.smegmatis_MC2_155 QKAAVYRDNAAWPGLAQAEQLHGKEMSYNNYTDADAAWRAAFDHEQTCVA
MAB_1064|M.abscessus_ATCC_1997 QQAALYIG-AAGQGLAQAEQLHGKEMSYNNYTDADAAWRAAWDQDKACAA
*:*::* . . ***********:*****:**********:*:: *.*
Mflv_1875|M.gilvum_PYR-GCK IIKHANPCGIAISSVSVADAHRKAHECDPLSAFGGVIAANTEVTVEMAET
Mvan_4856|M.vanbaalenii_PYR-1 IIKHANPCGIAISPVSVADAHRKAHECDPLSAFGGVIAANTEVTVEMAET
MUL_4712|M.ulcerans_Agy99 IIKHANPCGIAISEVSVADAHRKAHACDPLSAYGGVIACNTEVSPEMAEY
MMAR_4542|M.marinum_M IIKHANPCGIAISEVSVADAHRKAHACDPLSAYGGVIACNTEVSPEMAEY
Mb0982|M.bovis_AF2122/97 IIKHANPCGIAISSVSVADAHRKAHECDPLSAYGGVIAANTEVSVEMAEY
Rv0957|M.tuberculosis_H37Rv IIKHANPCGIAISSVSVADAHRKAHECDPLSAYGGVIAANTEVSVEMAEY
MLBr_00161|M.leprae_Br4923 IIKHANPCGIAISSLSVADAHRKAHECDPLSAYGGVIAANTEVSLEMAEY
MAV_1081|M.avium_104 IIKHANPCGIAISSTSVADAHRKAHECDPLSAFGGVIAANTEVSVEMAEY
MSMEG_5515|M.smegmatis_MC2_155 IIKHANPCGIAISSTSVADAHRKAHECDPLSAFGGVIAANTEVSVEMAET
MAB_1064|M.abscessus_ATCC_1997 IIKHANPCGIAVSDISIADAHLKAHECDPLSAFGGVIAVNREVSVEMAER
***********:* *:**** *** ******:***** * **: ****
Mflv_1875|M.gilvum_PYR-GCK VAGIFTEVIIAPAYEPGAVDVLKGKKNIRVLVASEPQPGGTEFRQISGGL
Mvan_4856|M.vanbaalenii_PYR-1 VAGIFTEVIIAPAYEPGAVEVLSGKKNIRVLVASEPQRGGTEFRQVSGGL
MUL_4712|M.ulcerans_Agy99 VSTIFTEVIVAPAYAPAALDQLTKKKNIRVLVASEPQDGGAELRPISGGL
MMAR_4542|M.marinum_M VSTIFTEVIVAPAYAPAALDQLTKKKNIRVLVASEPQDGGAELRPISGGL
Mb0982|M.bovis_AF2122/97 VSTIFTEVIVAPGYAPGALDVLARKKNIRVLVAAEPLAGGSELRPISGGL
Rv0957|M.tuberculosis_H37Rv VSTIFTEVIVAPGYAPGALDVLARKKNIRVLVAAEPLAGGSELRPISGGL
MLBr_00161|M.leprae_Br4923 VSTIFTEVIVAPSYAPGAVDVLSCKKNVRVLVASAPLRGGSELRPVSGGL
MAV_1081|M.avium_104 VSTIFTEVIIAPAYQPAALDILTRKKNIRVLVASEPLTGGTELRPISGGL
MSMEG_5515|M.smegmatis_MC2_155 ISEIFTEVIIAPAYEPGAVEVLARKKNIRILVASQPPIGGTEVRQISGGL
MAB_1064|M.abscessus_ATCC_1997 VADIFTEVIVAPGYADGAVEVLSRKKNVRLLRAAQPESGSTEWRQISGGL
:: ******:**.* .*:: * ***:*:* *: * *.:* * :****
Mflv_1875|M.gilvum_PYR-GCK LLQQRDALDASGDDPNNWTLATGAPADPDTLADLVFAWRTCRAVKSNAIV
Mvan_4856|M.vanbaalenii_PYR-1 LLQQRDALDAAGDNPNTWTLAAGPAADPDTLADLAFAWRTCRAVKSNAIV
MUL_4712|M.ulcerans_Agy99 LMQQRDQLDAAGDNPANWILATGSPADPATLTDLVFAWRSCRAVKSNAIV
MMAR_4542|M.marinum_M LMQQRDQLDAAGDNPANWTLATGSPADPATLTDLVFAWRSCRAVKSNAIV
Mb0982|M.bovis_AF2122/97 LIQQSDQLDAHGDNPANWTLATGSPADPATLTDLVFAWRACRAVKSNAIV
Rv0957|M.tuberculosis_H37Rv LIQQSDQLDAHGDNPANWTLATGSPADPATLTDLVFAWRACRAVKSNAIV
MLBr_00161|M.leprae_Br4923 LIQQPDQLDTAGDNPANWTLATGSPAGPATLTDLVFAWRACRAVKSNAIV
MAV_1081|M.avium_104 LVQQRDELDAHGDNPANWTLATGAPADPATLADLVFAWRVCRAVKSNAIV
MSMEG_5515|M.smegmatis_MC2_155 LLQQRDALDADGDDPNNWTLATGEPADPKTLEDLVFAWRACRAVKSNAIV
MAB_1064|M.abscessus_ATCC_1997 LVQERDALDAEGDDPANWTLVAGAAADADTLADLVFAWKVGRAVKSNAIV
*:*: * **: **:* .* *.:* .*.. ** **.***: *********
Mflv_1875|M.gilvum_PYR-GCK LAKDGATVGVGMGQVNRVDAARLAVERAGERTRDAVGASDAFFPFPDGLQ
Mvan_4856|M.vanbaalenii_PYR-1 LARDGATVGVGMGQVNRVDAARLAVERAGGRSSGAVGASDAFFPFPDGLE
MUL_4712|M.ulcerans_Agy99 IVADGATIGVGMGQVNRVDAARLAVERGGERVNGAVAASDAFFPFPDGLE
MMAR_4542|M.marinum_M IVADGATIGVGMGQVNRVDAARLAVERGGERVNGAVAASDAFFPFPDGLE
Mb0982|M.bovis_AF2122/97 IAADGATVGVGMGQVNRVDAARLAVERGGERVRGAVAASDAFFPFPDGLE
Rv0957|M.tuberculosis_H37Rv IAADGATVGVGMGQVNRVDAARLAVERGGERVRGAVAASDAFFPFPDGLE
MLBr_00161|M.leprae_Br4923 IAADGATIGVGMGQVNRVDAARLAVERGGDRVRGAVVASDAFFPFADGLQ
MAV_1081|M.avium_104 IAAGGATIGVGMGQVNRVDAARLAVERGGDRVRGAVAASDAFFPFPDGLE
MSMEG_5515|M.smegmatis_MC2_155 VARDGATVGVGMGQVNRVDAAKLAVQRGGDRVAGAVAASDAFFPFPDGLQ
MAB_1064|M.abscessus_ATCC_1997 IASGGATIGVGMGQVNRVDAARLAVSRGGDRVSGAVAASDAFFPFPDGLE
:. .***:*************:***.*.* * .** ********.***:
Mflv_1875|M.gilvum_PYR-GCK TLIDAGVKAVVHPGGSVRDDEVTAAAEAAGITLYLTGARHFAH
Mvan_4856|M.vanbaalenii_PYR-1 TLIRAGVKAVVHPGGSVRDDEVTAAAEADGITLYLTGARHFAH
MUL_4712|M.ulcerans_Agy99 TLTAAGVKAIVHPGGSVRDEEVTAAAAKAGITLYLTGSRHFVH
MMAR_4542|M.marinum_M TLTAAGVKAIVHPGGSVRDEEVTAAAAKAGITLYLTGSRHFAH
Mb0982|M.bovis_AF2122/97 TLAAAGVTAVVHPGGSVRDEEVTEAAAKAGVTLYLTGARHFAH
Rv0957|M.tuberculosis_H37Rv TLAAAGVTAVVHPGGSVRDEEVTEAAAKAGVTLYLTGARHFAH
MLBr_00161|M.leprae_Br4923 TLAAAGVTAIVHPGGSVRDAEVTAAATKAGVTLYLTGVRHFVH
MAV_1081|M.avium_104 TLTGAGVKAVVHPGGSVRDDEVTAAAANAGITLYLTGARHFAH
MSMEG_5515|M.smegmatis_MC2_155 VLTEADVKAIVHPGGSVRDAEVTAAAELAGITLYLTGARHFAH
MAB_1064|M.abscessus_ATCC_1997 VLSQAGVRAVVHPGGSMRDELVTDAAKAAGITLYTTGARHFAH
.* *.* *:******:** ** ** *:*** ** ***.*