For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLTAAAALVLTLTPGCSSSPTPSANAANPQRRIDTVTPPGLRAQQTMDMLNSDWPIGPAGVATLAAPSKV DSVEATMEGLWWDRPFALDSVDISASVATLHLTSSYGARQDIRLHTDDGGWVDRFELATLPPPTITAWRD VDAVLSKTGARYSYQVSKVEKGQCTRVAGTNTNQPLPLASIFKLYVLHALADAIKSGTASWDDQLTVTAK SKAVGSSGLELSPGDHVSVRRAAEKMIANSDNMATDLLIGRLGKHAIEQALATAGHHDPASMTPFPTMYE LFSVGWGQPDLREQWKQASAQQRAALLEQADSTPYRPDPTRAHTPASNYGAEWYGSAEDICRVHLALQAD AVGPAAPVRKILSAVAGIALDRNYWPYIGAKAGGLPGDLTFSWYAEDKTGQPWVVSFQLNWPRDHGPNVT GWMLQVAKQVFALVAPE
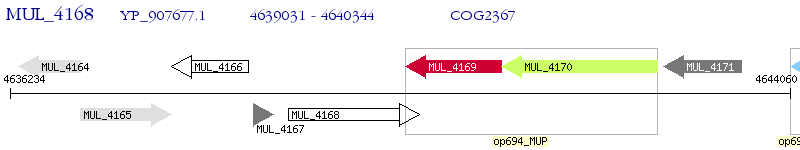
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4168 | lpqF | - | 100% (437) | lipoprotein LpqF |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3624 | lpqF | 0.0 | 78.90% (436) | lipoprotein LpqF |
| M. gilvum PYR-GCK | Mflv_1434 | - | 1e-153 | 61.52% (434) | beta-lactamase |
| M. tuberculosis H37Rv | Rv3593 | lpqF | 0.0 | 78.90% (436) | lipoprotein LpqF |
| M. leprae Br4923 | MLBr_01923 | lpqF | 0.0 | 76.78% (435) | probale secreted protein |
| M. abscessus ATCC 19977 | MAB_0552c | - | 1e-144 | 60.14% (429) | lipoprotein LpqF |
| M. marinum M | MMAR_5092 | lpqF | 0.0 | 99.54% (437) | lipoprotein LpqF |
| M. avium 104 | MAV_0561 | - | 0.0 | 78.26% (437) | LpqF protein |
| M. smegmatis MC2 155 | MSMEG_6087 | - | 1e-160 | 63.51% (433) | beta-lactamase |
| M. thermoresistible (build 8) | TH_0594 | lpqF | 1e-157 | 62.56% (438) | PROBABLE CONSERVED LIPOPROTEIN LPQF |
| M. vanbaalenii PYR-1 | Mvan_5353 | - | 1e-153 | 62.53% (435) | beta-lactamase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1434|M.gilvum_PYR-GCK -MRGLTPTPEAARARR-GATVVVALLSVVALACGCAATRPAPADAAY-GA
Mvan_5353|M.vanbaalenii_PYR-1 -MRGLTPRRYAGRARRRATTAAVALVCVASLACGCAATRPAPADAAY-GA
MSMEG_6087|M.smegmatis_MC2_155 ------------------------MAVVAALACGCSERSAPAAEVSY-GA
TH_0594|M.thermoresistible__bu -LPEAAPTSNARRSRR-LIGLLAAAVLTAASAGACADRGTPPADAAY-GT
MUL_4168|M.ulcerans_Agy99 -------------------MLTAAAALVLTLTPGCSSSPTPSANAANPQR
MMAR_5092|M.marinum_M MQGTRRLTSRRRNRLRRATVLTAAAAMVLTLTPGCSSSPTPSANAANPQR
MLBr_01923|M.leprae_Br4923 MPQPARRTNQRPPRH-RTVALAATAALVVTMAPGCAHSASPYANTADPGL
MAV_0561|M.avium_104 -------------------MLSAAAALVATLAPGCASSPSPQANAANPGR
Mb3624|M.bovis_AF2122/97 -MGPARLHNRRAGR--RMLALSAAAALIVALASGCSSAPTPSANAANHGH
Rv3593|M.tuberculosis_H37Rv -MGPARLHNRRAGR--RMLALSAAAALIVALASGCSSAPTPSANAANHGH
MAB_0552c|M.abscessus_ATCC_199 -------MNEGSLRARMTRAVSVCTVAAVTLALGGTAGCGIKLRQVP-HG
: : . :
Mflv_1434|M.gilvum_PYR-GCK PIQINTPQGLRAKQTMDMLNSDWPIGEVGVRTLAAPDLVEHVGVTLDRIW
Mvan_5353|M.vanbaalenii_PYR-1 PIQINTPQGLRAKQTMDMLNSDWPIGEVGVRTLAAPEAVEHVGVTLDRIW
MSMEG_6087|M.smegmatis_MC2_155 HIDTNTPPGLRAKQTMDMLNSDWPIGPVSVGTLASPEKIDDVGVKMDTIW
TH_0594|M.thermoresistible__bu HIPTGTPQGLRAKQTMDMLNSDWPIGEPSVSTLAAPEHVAEVTESMETLW
MUL_4168|M.ulcerans_Agy99 RIDTVTPPGLRAQQTMDMLNSDWPIGPAGVATLAAPSKVDSVEATMEGLW
MMAR_5092|M.marinum_M RIDTVTPPGLRAQQTMDMLNSDWPIGPAGVATLAAPSKVDSVEATMEGLW
MLBr_01923|M.leprae_Br4923 RIDTRTPPGVRAQQIIDMLNSDWPIGLVGVRTFASHDMVEIVKTTLEELW
MAV_0561|M.avium_104 QIDTRTPPGIRAQQTLDMLNSDWPIGPVGVRTLAAADKVESVQTTLEALW
Mb3624|M.bovis_AF2122/97 RIDTRTPPGLRAQQTMDMLNSDWPIGEIGVGTLAAPGQVDTVKTTMEALW
Rv3593|M.tuberculosis_H37Rv RIDTRTPPGLRAQQTMDMLNSDWPIGEIGVGTLAAPGQVDTVKTTMEALW
MAB_0552c|M.abscessus_ATCC_199 DVDTSTPPGLRSQQLIDMLNSNWPIGKQGVATMAAANKVDDYTEIMTKLW
: ** *:*::* :*****:**** .* *:*: : : :*
Mflv_1434|M.gilvum_PYR-GCK WDRPFTLTDVDVGAGWATLDVQTSYGVAQTIRLRTDEQGLVDRFSVDLVP
Mvan_5353|M.vanbaalenii_PYR-1 WDRPFTVTNVDVGAGHAKLDVLTSYGVAQTIELRTDEAGMVDRFQVQLVP
MSMEG_6087|M.smegmatis_MC2_155 WDRPFKVTSVEIGAGQATLHVLTSYNVAQNIELRTNDAGLVDRFEVNLEP
TH_0594|M.thermoresistible__bu WDRPITLTAVDIGAGTATLRVRNSYGAEQDIELRVDDESLVDQFEVTLRP
MUL_4168|M.ulcerans_Agy99 WDRPFALDSVDISASVATLHLTSSYGARQDIRLHTDDGGWVDRFELATLP
MMAR_5092|M.marinum_M WDRPFALDSVDISASVATLHLTSSYGARQDIRLHTDDGGWVDRFELETLP
MLBr_01923|M.leprae_Br4923 WDRPFTLEGVDIGASVATLHLISSYGAQQDIRIHTDGNSWVDRFDLNTQP
MAV_0561|M.avium_104 WDRPFTLNGVDIGASVATLHLTSSYGARQDIRIHTDDGGWVDRFDLDTRA
Mb3624|M.bovis_AF2122/97 WDRPFALAGVDIGASVAALHLISSYGAQQDIRIHTDDDGWVDRFDVETQA
Rv3593|M.tuberculosis_H37Rv WDRPFALAGVDIGASVAALHLISSYGAQQDIRIHTDDDGWVDRFDVETQA
MAB_0552c|M.abscessus_ATCC_199 VDRPYKVSSVDLGANSSTVHLIAPYGANVDIGLRTNDKGDVDRLVPYQQP
*** : *::.*. : : : .*.. * ::.: . **:: .
Mflv_1434|M.gilvum_PYR-GCK P-EIDEWSDIDEQLTASGARYSYQVAKVDD-----GRCVKVAGSNTDLSL
Mvan_5353|M.vanbaalenii_PYR-1 P-KIATWSDIDEQLTKSGARYSYQVAKVTD-----GQCVKVAGTNTDTSL
MSMEG_6087|M.smegmatis_MC2_155 P-VINGWSDIDKEIAKTGARYSYQVSKVRPGAEATDACEVVAGTNTDLSL
TH_0594|M.thermoresistible__bu P-TIENWADIDTELTESGARYSYQVSRVLP-APGGRRCEPIAGANTDLSL
MUL_4168|M.ulcerans_Agy99 PPTITAWRDVDAVLSKTGARYSYQVSKVE-----KGQCTRVAGTNTNQPL
MMAR_5092|M.marinum_M PPTITAWRDVDAVLSKTGARYSYQVSKVE-----KGQCTRVAGTNTNQPL
MLBr_01923|M.leprae_Br4923 P-SITSWYDIDAALSKTGARYSYQVAKVD-----NGRCDPVQGTNTAESL
MAV_0561|M.avium_104 P-TVNSWHDVDAVLSKTGARYSYQAAKVDP----QGRCEPVAGTNTGESL
Mb3624|M.bovis_AF2122/97 P-SIASWRDVDAVLSKTGARYSFQVAKVD-----NGRCDPVAGTSTGESL
Rv3593|M.tuberculosis_H37Rv P-SIASWRDVDAALSKTGARYSFQVAKVD-----NGRCDPVAGTNTGESL
MAB_0552c|M.abscessus_ATCC_199 P-TIANWSDVDAALSASGGRYSYRASKIVD-----GQCQQIAGTNTSQAM
* : * *:* :: :*.***::.::: * : *:.* .:
Mflv_1434|M.gilvum_PYR-GCK PLASIFKLYVLLAVADAVKAGTLQWSDPLTITEEAKAVGSSGFDELEPGA
Mvan_5353|M.vanbaalenii_PYR-1 PLASIFKLYVLFAVAEAVKADTLEWTDALTITEGAKAVGSSAFDELEPGA
MSMEG_6087|M.smegmatis_MC2_155 PLASIFKLYVLLAVSDAIKEGTLSWDDKLTITKDGKALGSAGLDKLPPGA
TH_0594|M.thermoresistible__bu PLASIFKLYVLLAVAEAVRAGTLDWDDRLTVTERAKAVGSAALDHLPPGA
MUL_4168|M.ulcerans_Agy99 PLASIFKLYVLHALADAIKSGTASWDDQLTVTAKSKAVGSSGL-ELSPGD
MMAR_5092|M.marinum_M PLASIFKLYVLHALADAIKSGTASWDDQLTVTAKSKAVGSSGL-ELSPGD
MLBr_01923|M.leprae_Br4923 PLASIFKLYVLHALAYAVRNGTVSWDDQLTVTDKSRAVGSSGL-ELPIGE
MAV_0561|M.avium_104 PLASIFKLYVLHALAGAVTAGRVSWDDQLTVTDKSRAVGSSGL-ALPAGA
Mb3624|M.bovis_AF2122/97 PLASIFKLYVLHALAGAVQHNTVSWDDLLTVTAKSKAVGSSGL-ELPVGA
Rv3593|M.tuberculosis_H37Rv PLASIFKLYVLHALAGAVQHNTVSWDDLLTVTAKSKAVGSSGL-ELPVGA
MAB_0552c|M.abscessus_ATCC_199 PLASVFKLYVLLAAGTAINAGTLRWDDQLTVTDRGKALG-SSMDKLPTGS
****:****** * . *: . * * **:* .:*:* :.: * *
Mflv_1434|M.gilvum_PYR-GCK TVTVREASQQMISASDNMATDLLIERLGPGAIERALAEAGHHDPASMTPF
Mvan_5353|M.vanbaalenii_PYR-1 TVSVREAAQQMISASDNMATDLLIERLGPGAIERALVAAGHHDPASMTPF
MSMEG_6087|M.smegmatis_MC2_155 QITVRTAAQQMISASDNMATDLLIGRVGTGAVERALVTAGHHDPASMTPF
TH_0594|M.thermoresistible__bu QISVREAAQQMISASDNMTTDLLIGRLGTDAVERALVAAGHHDPASMTPF
MUL_4168|M.ulcerans_Agy99 HVSVRRAAEKMIANSDNMATDLLIGRLGKHAIEQALATAGHHDPASMTPF
MMAR_5092|M.marinum_M HVSVRRAAEKMIANSDNMATDLLIGRLGKHAIEQALATAGHHDPASMTPF
MLBr_01923|M.leprae_Br4923 HVSVRTAAEKMIATSDNMATDLLIGKMGTHAVEEALVTAGHHDPASMTPF
MAV_0561|M.avium_104 QVSVRTAAEKMIATSDNMATDLLIDKLGTHAIEEALATAGHHDPSSMTPF
Mb3624|M.bovis_AF2122/97 RVSVRTAAEKMIATSDNMATDLLIERLGTRAIEEALASAGHHDPASMTPF
Rv3593|M.tuberculosis_H37Rv RVSVRTAAEKMIATSDNMATDLLIERLGTRAIEEALASAGHHDPASMTPF
MAB_0552c|M.abscessus_ATCC_199 TVSVRTAAQKMISVSDNMGTDMLINRVGRHAIEKALADAGHHDPASMTPF
::** *:::**: **** **:** ::* *:*.**. ******:*****
Mflv_1434|M.gilvum_PYR-GCK PTMHELFSVGWGTPDLREQWKKAS-PEGRAQLLQQTNSRPYEPDPKRTHM
Mvan_5353|M.vanbaalenii_PYR-1 PTMHELFSVGWGTPDLREQWKRAT-REGRAQLLQQTNSRPYEPDPKRTHL
MSMEG_6087|M.smegmatis_MC2_155 PTMHQVFSVGWGEPNLREQWKNAS-REDRIKLLQQTDSRPYQPDPNRTHS
TH_0594|M.thermoresistible__bu PTMHELFSVGWGEPDLREQWQHAS-PQDRAELLRQVSSRPYRPDPQRTHT
MUL_4168|M.ulcerans_Agy99 PTMYELFSVGWGQPDLREQWKQAS-AQQRAALLEQADSTPYRPDPTRAHT
MMAR_5092|M.marinum_M PTMYELFSVGWGQPDLREQWKQAS-AQQRAALLEQADSTPYRPDPTRAHT
MLBr_01923|M.leprae_Br4923 PTMYELFSVGWGQPDLRSQWKQAS-QQLRAQLLQQANSTPYQPDPTRAHT
MAV_0561|M.avium_104 PTMYELFSVGWGRPDLRSQWEHGS-PQVRARLLAQANSTPYDPDPMRAHS
Mb3624|M.bovis_AF2122/97 PTMYELFSVGWGKPDLRDQWKHAT-QQVRAQILRQTNSTPYQPDPTRAHT
Rv3593|M.tuberculosis_H37Rv PTMYELFSVGWGKPDLRDQWKHAT-QQVRAQILRQTNSTPYQPDPTRAHT
MAB_0552c|M.abscessus_ATCC_199 PTMHELFNIGWGIPDVRQQWKDATTPEQRGRMLAEADTHDYKLDPDRTTT
***:::*.:*** *::*.**: .: : * :* :..: * ** *:
Mflv_1434|M.gilvum_PYR-GCK PASEYGAEWYGSAADICRLHAALQRSAVGEAAPVKDILSFLPGIDLDRAE
Mvan_5353|M.vanbaalenii_PYR-1 PASDIGAEWYGSAADICRLHAALQKGAVGEAAPVRDILSAVPGIDLDRDE
MSMEG_6087|M.smegmatis_MC2_155 PGSTDGLEWYGSAQDICRVHAALQASAVGPAAPVKDILSALPGIDLDPAK
TH_0594|M.thermoresistible__bu PASNYGAEWFGTAKDICRVHIALQEAAVGPAAPVRDILSAIPGVSVDRST
MUL_4168|M.ulcerans_Agy99 PASNYGAEWYGSAEDICRVHLALQADAVGPAAPVRKILSAVAGIALDRNY
MMAR_5092|M.marinum_M PASNYGAEWYGSAEDICRVHLALQADAVGPAAPVRKILSAVAGIALDRNY
MLBr_01923|M.leprae_Br4923 PASNYGAEWYGSAEDICRIHAALQADAIGQAAPVRQILSAVAGIQLDRSK
MAV_0561|M.avium_104 PASSYGAEWYGNAQDICRVHAALQADAVGKAAPVRDILSAVSGIQLDPAV
Mb3624|M.bovis_AF2122/97 PASNYGAEWYGSAEDICRVHAALRADAVGPASPVRQIMSAVPGIQLDRSV
Rv3593|M.tuberculosis_H37Rv PASNYGAEWYGSAEDICRVHAALRADAVGPASPVRQIMSAVPGIQLDRSV
MAB_0552c|M.abscessus_ATCC_199 PASKYGIEWYGSAEDICRVHAALQKVAVGPAAPVRDILAAEPGIDLHSEN
*.* * **:*.* ****:* **: *:* *:**:.*:: .*: :.
Mflv_1434|M.gilvum_PYR-GCK WPYIGAKAGNLPGDMTFSWYAVDRSGQPWVVSLQSNWPEFRSQTAAAWLM
Mvan_5353|M.vanbaalenii_PYR-1 WPYIGAKAGNLPGDLTFSWYAVDASGQPWVVSLQSNWPEFRSQTAAAWLM
MSMEG_6087|M.smegmatis_MC2_155 WTYIGAKGGNLPGDLTFSWYAVDHTGQPWVVSFQLNWPQFRSSTAAGWLL
TH_0594|M.thermoresistible__bu WTYIAAKGGNLPGDLTFSWYAVDHTGQPWVVSFQLNWPQYRSKTAAGWLL
MUL_4168|M.ulcerans_Agy99 WPYIGAKAGGLPGDLTFSWYAEDKTGQPWVVSFQLNWPRDHGPNVTGWML
MMAR_5092|M.marinum_M WPYIGAKAGGLPGDLTFSWYAEDKTGQPWVVSFQLNWPRDHGPNVTGWML
MLBr_01923|M.leprae_Br4923 WPYIGAKAGGLPGDLTFSWYAVDKTQQSWVVSFQLNWPRDHGPTVTSWML
MAV_0561|M.avium_104 WPYIGAKAGGLPGDLTFSWYAVDKTHQPWVVSFQLNWPRDHGPTATGWML
Mb3624|M.bovis_AF2122/97 WPYIGAKAGGLPGDLTFSWYAVDKTGQPWVVSFQLNWPRDHGPTVTGWML
Rv3593|M.tuberculosis_H37Rv WPYIGAKAGGLPGDLTFSWYAVDKTGQPWVVSFQLNWPRDHGPTVTGWML
MAB_0552c|M.abscessus_ATCC_199 WRYIGAKGGNLPGDLTFSWYAEDRTRQPYVVSFQLNWDRAFGPGAAGWVI
* **.**.*.****:****** * : *.:***:* ** . . .:.*::
Mflv_1434|M.gilvum_PYR-GCK SIARQTFDLIPN----
Mvan_5353|M.vanbaalenii_PYR-1 SIARQTFDLVPNG---
MSMEG_6087|M.smegmatis_MC2_155 QIAKRAFALAPVGRLH
TH_0594|M.thermoresistible__bu SIVHQAFELIPVG---
MUL_4168|M.ulcerans_Agy99 QVAKQVFALVAPE---
MMAR_5092|M.marinum_M QVAKQVFALVAPE---
MLBr_01923|M.leprae_Br4923 QLAKQVFALFAPR---
MAV_0561|M.avium_104 QVAKQAFALIAPR---
Mb3624|M.bovis_AF2122/97 QVARQVFALIAPQ---
Rv3593|M.tuberculosis_H37Rv QVARQVFALIAPQ---
MAB_0552c|M.abscessus_ATCC_199 GLAKGVFKKLG-----
:.: .*