For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTAALNQRPRGGAPAPTRSRDFWGSATRLVKRLAPQRRRSLAVIALGIVGTAIGVAVPRILGHAADLLFN GVIGRQLPAGITKAQAIAGARARGDNTFAELLSGMKVTPGQGVDFGAVVRTLALALALYLIASLLIWGQA RLLNVTVQRTVAALRADVEHKLHRLPLSYFDGRQRGELLSRVTNDIDNIQSSLSMTISQLVTAILTVVAV LSMMVWISPLLALITLLTVPLSLLVTRAITRRSQRLFVAHWTSTGRLNAHIEETYSGFTVVKTFGHQAAA REQFHNLNDDVYQASFGAQLLSALVQPATTFIGNLGYVAVAVMGGLQVATGQITLGSVQAFIQYVRQFNT PVSQVAAMYNSLQSGVASAERVFELLDEPEEAADPASALQPPVSRTGRAPALTGRVEFERVNFAYHPGHP VIHDLSLVAEPGSTVAIVGPTGAGKTTLVNLLMRFYEVDSGRILVDGIDITTVSRQSLRSHIGMVLQDTW LFDGTISENIAYGRPEASPDDIIQAAKAAYVDRFVHMLPASYQTRISGDGSNISAGEKQLITIARAFLAR PQLLILDEATSSVDTRTELLIARATAELRRDRTSFIIAHRLSTIRDADQILVVQAGQIVEHGTHAELLAN RGAYYAMTRA
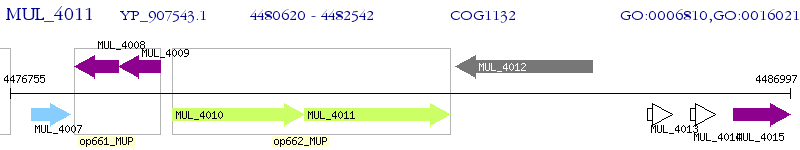
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4011 | - | - | 100% (640) | transmembrane ATP-binding protein ABC transporter |
| M. ulcerans Agy99 | MUL_3902 | - | 1e-68 | 35.00% (540) | transmembrane ATP-binding protein ABC transporter |
| M. ulcerans Agy99 | MUL_4010 | - | 6e-66 | 33.12% (459) | transmembrane ATP-binding protein ABC transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1303c | - | 0.0 | 81.59% (641) | drugs-transport transmembrane ATP-binding protein ABC |
| M. gilvum PYR-GCK | Mflv_2250 | - | 0.0 | 73.57% (628) | ABC transporter, transmembrane region |
| M. tuberculosis H37Rv | Rv1272c | - | 0.0 | 81.59% (641) | drugs-transport transmembrane ATP-binding protein ABC |
| M. leprae Br4923 | MLBr_01113 | - | 0.0 | 75.64% (628) | putative ABC transporter, ATP-binding component |
| M. abscessus ATCC 19977 | MAB_1414c | - | 0.0 | 70.47% (640) | ABC transporter ATP-binding protein |
| M. marinum M | MMAR_4148 | - | 0.0 | 99.38% (640) | transmembrane ATP-binding protein ABC transporter |
| M. avium 104 | MAV_1420 | - | 0.0 | 77.09% (633) | ABC transporter |
| M. smegmatis MC2 155 | MSMEG_5009 | - | 0.0 | 73.42% (632) | ABC transporter |
| M. thermoresistible (build 8) | TH_3535 | - | 0.0 | 73.32% (641) | PUTATIVE ABC transporter, transmembrane region |
| M. vanbaalenii PYR-1 | Mvan_4441 | - | 0.0 | 73.08% (639) | ABC transporter, transmembrane region |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2250|M.gilvum_PYR-GCK MSGPLARGIR-MGEPPQERSRDFKASAIRLLRRLTPQRGMTIAVMLLGVG
Mvan_4441|M.vanbaalenii_PYR-1 MTGPLARSMR-MAEPPQTRSRDFTGSALRLFRRLTPQRGMTVAVMLLGVG
MSMEG_5009|M.smegmatis_MC2_155 -----MRRGA-LPQAPLERTRDFKGSAIRLARRLLPQRALTLAVILLGVG
TH_3535|M.thermoresistible__bu VTGPMNRPLRGVVQAPTERSSDFTGTALRLVRRLAPQRGPALLVVLLGIG
MUL_4011|M.ulcerans_Agy99 MTAALNQRPRGGAPAP-TRSRDFWGSATRLVKRLAPQRRRSLAVIALGIV
MMAR_4148|M.marinum_M MTAALNQRPRGGAPAP-TRSRDFWGSATRLVKRLAPQRRRSLAVIALGIV
Mb1303c|M.bovis_AF2122/97 MTAPPGARPRAASPPPNMRSRDFWGSAARLVKRLAPQRRLSIAVITLGIA
Rv1272c|M.tuberculosis_H37Rv MTAPPGARPRAASPPPNMRSRDFWGSAARLVKRLAPQRRLSIAVITLGIA
MLBr_01113|M.leprae_Br4923 MTMAT---------APATRSCDFYCTIRRLLKQLVPQRRLSLTVITLGIT
MAV_1420|M.avium_104 MTTTTRPAP----PAPAGRSRDFWGSAARLVKQLAPQRRTAIAVITLGIA
MAB_1414c|M.abscessus_ATCC_199 MTRAIGFRG--VQQGPAERSRDFRGTAIRMVKRLAPQRFLTATVITLSMI
* *: ** : *: ::* *** : *: *.:
Mflv_2250|M.gilvum_PYR-GCK GIAIGVVGPRILGHATDLLFNGVIGRQLPAGSTKEQAIEAARARGDTTFA
Mvan_4441|M.vanbaalenii_PYR-1 GIAIGVIGPRILGHATDLLFNGVIGRQLPAGLTKEQAVEAARARGDGTFA
MSMEG_5009|M.smegmatis_MC2_155 GIAIGVIGPRILGHATDLLFNGVIGRELPAGLTKEQAVEAARARGDGTFA
TH_3535|M.thermoresistible__bu GIAIGVIGPRILGHATDLLFNGVIGRQLPAGITKEQAVEAARARGDSTFA
MUL_4011|M.ulcerans_Agy99 GTAIGVAVPRILGHAADLLFNGVIGRQLPAGITKAQAIAGARARGDNTFA
MMAR_4148|M.marinum_M GTAIGVAVPRILGHATDLLFNGVIGRQLPAGITKAQAIAGARARGDNTFA
Mb1303c|M.bovis_AF2122/97 GTTIGVIVPRILGHATDLLFNGVIGRGLPGGITKAQAVASARARGDNTFA
Rv1272c|M.tuberculosis_H37Rv GTTIGVIVPRILGHATDLLFNGVIGRGLPGGITKAQAVASARARGDNTFA
MLBr_01113|M.leprae_Br4923 GTAIGIIVPWVLGHTTDLLFNGVIGRQLPKGISKTQAVAAARTRGDNALA
MAV_1420|M.avium_104 GTVIGVMVPRILGHATDLLFNGVVGRQLPAGISKAQAVAAARSRGQNTFA
MAB_1414c|M.abscessus_ATCC_199 GIAIGVIGPRILGHATDLLFNGVIGRQLPAGQSREQAVETARARGDGQFA
* .**: * :***::*******:** ** * :: **: **:**: :*
Mflv_2250|M.gilvum_PYR-GCK DLLSGMDVVPGQGVDFAAVARTLMLALGLYLVAALMVWIQARLLNVVVQR
Mvan_4441|M.vanbaalenii_PYR-1 DLLSGMDVVPGQGVDFGAVGRTLALALALYLVAALMVWLQALLLNIVVQR
MSMEG_5009|M.smegmatis_MC2_155 DLLSGMDIVPGQGVDFGAVGRTLALALGLYLVAALLVWVQARLLNVTVQR
TH_3535|M.thermoresistible__bu DMVSAMDVVPGQGVDFGAVARTLLVALGLYLLAAVLVWLQFRVLNVTVQR
MUL_4011|M.ulcerans_Agy99 ELLSGMKVTPGQGVDFGAVVRTLALALALYLIASLLIWGQARLLNVTVQR
MMAR_4148|M.marinum_M ELLSGMKVTPGQGVDFGAVVRTLALALALYLIASLLIWGQARLLNVTVQR
Mb1303c|M.bovis_AF2122/97 DLLSGMNVVPGQGVDFAAVERTLALALALYLAAALMIWAQARLLNLTVQK
Rv1272c|M.tuberculosis_H37Rv DLLSGMNVVPGQGVDFAAVERTLALALALYLAAALMIWAQARLLNLTVQK
MLBr_01113|M.leprae_Br4923 DLLSGMNLVPGQGVDFVAVGRTLGLALSLYLVYTLLTWAQARLLNVTVQR
MAV_1420|M.avium_104 DLLSGMNVVPGRGVDFGAVARTLALALLLYLVSALLIWVQARLLNRTVQR
MAB_1414c|M.abscessus_ATCC_199 QMLSGMNVIPGQGVDFHAIGMTLALALSLYLIAGMLAWVQARLLNVTVQR
:::*.*.: **:**** *: ** :** *** :: * * :** .**:
Mflv_2250|M.gilvum_PYR-GCK TMVSLRADVEDKVHRLPLRYFDSRQRGEVLSRVTNDVDNLQQSLTMSITQ
Mvan_4441|M.vanbaalenii_PYR-1 TMVALRADVEDKVHRLPLRYFDSRQRGEVLSRVTNDVDNLQQSLSMSITQ
MSMEG_5009|M.smegmatis_MC2_155 TMVALRAEVQEKIHRLPLSYFDSRQRGEVLSRVTNDVDNIQNSVSMTISQ
TH_3535|M.thermoresistible__bu TIMTLRAEVEDKVHRLPLSYFDSRRRGEVLSRVTNDVDNIATSLSMTISQ
MUL_4011|M.ulcerans_Agy99 TVAALRADVEHKLHRLPLSYFDGRQRGELLSRVTNDIDNIQSSLSMTISQ
MMAR_4148|M.marinum_M TVAALRADVEHKLHRLPLSYFDGRQRGELLSRVTNDIDNIQSSLSMTISQ
Mb1303c|M.bovis_AF2122/97 TMVRLRTDVEDKVHRLPLSYFDGQQRGELLSRVTNDIDNLQSSLSMTISQ
Rv1272c|M.tuberculosis_H37Rv TMVRLRTDVEDKVHRLPLSYFDGQQRGELLSRVTNDIDNLQSSLSMTISQ
MLBr_01113|M.leprae_Br4923 IMFALRSDVEDKVHRLPLSYCDTRQRGELLSRVTNDIDNLQSSLSITLSQ
MAV_1420|M.avium_104 TMVALRSAVEDKVHRLPLAYFDGRQRGELLSRVTNDIDNIQSSLSMTISQ
MAB_1414c|M.abscessus_ATCC_199 TVVALRTEVENKIHRLPLSYFDSRQRGEILSRVTNDVDNIQTSLQMSINQ
: **: *:.*:***** * * ::***:*******:**: *: :::.*
Mflv_2250|M.gilvum_PYR-GCK LLTSVLTVVAVLVMMLTISPMLTLLTVVTVPLSLWVTRSIAKRSRKLFVA
Mvan_4441|M.vanbaalenii_PYR-1 LLTSVLTVIAVLVMMLTISPLLTLLTVVTVPLSLWVTRSIAKRSRTLFVA
MSMEG_5009|M.smegmatis_MC2_155 LLTSVLTVFAVLVMMLTISPLLTLFTVVTVPASLWVTRWITRRSQPLFVA
TH_3535|M.thermoresistible__bu LLTSLLTLVAVLVMMVTISPLLALITVLTVPLSLWATRTIARRSQRMFVA
MUL_4011|M.ulcerans_Agy99 LVTAILTVVAVLSMMVWISPLLALITLLTVPLSLLVTRAITRRSQRLFVA
MMAR_4148|M.marinum_M LVTAILTVVAVLSMMVWISPLLALITLLTVPLSLLVTRAITRRSQRLFVA
Mb1303c|M.bovis_AF2122/97 LVTSILTMVAVLAMMVSISGLLALITLLTVPLSLLVTRAITRRSQPLFVA
Rv1272c|M.tuberculosis_H37Rv LVTSILTMVAVLAMMVSISGLLALITLLTVPLSLLVTRAITRRSQPLFVA
MLBr_01113|M.leprae_Br4923 LLTSLLTVVAVLVVMVSVSPLLALVALLTVPMSLLATRAIAQRSQQLFVA
MAV_1420|M.avium_104 LLTSVLTVLTVLAMMLSISPLLAGITLLTVPLSLLATRVIVRRSQRLFVA
MAB_1414c|M.abscessus_ATCC_199 LLSSMLTLVAVLAMMLSISPLLAVITLATVPLSLWVTRTIARRSQRLFVA
*::::**:.:** :*: :* :*: .:: *** ** .** *.:**: :***
Mflv_2250|M.gilvum_PYR-GCK QWTNTGRLNAHIEETYSGFTVVKTFGHRARAEDQFRELNDDVYHASFGAQ
Mvan_4441|M.vanbaalenii_PYR-1 QWTNTGRLNAHIEETYSGFTVVKTFGHRAHAEDQFRQLNDDVYHASFGAQ
MSMEG_5009|M.smegmatis_MC2_155 QWRNTGRLAAHLEETYSGFTIVKTFGHREAAAGKFAELNSETQQSSFGAQ
TH_3535|M.thermoresistible__bu QWRNTGRLNAHIEETYSGFTVVKTYGHRAHAEDQFRALNAEVYRSGFGAQ
MUL_4011|M.ulcerans_Agy99 HWTSTGRLNAHIEETYSGFTVVKTFGHQAAAREQFHNLNDDVYQASFGAQ
MMAR_4148|M.marinum_M HWTSTGRLNAHIEETYSGFTVVKTFGHQAAAREQFHNLNDDVYQASFGAQ
Mb1303c|M.bovis_AF2122/97 HWTSTGRLNAHLEETYSGFTVVKTFGHQAAARERFHELNDDVYQAGFGAQ
Rv1272c|M.tuberculosis_H37Rv HWTSTGRLNAHLEETYSGFTVVKTFGHQAAARERFHELNDDVYQAGFGAQ
MLBr_01113|M.leprae_Br4923 QWTSAGRLNAHIEETYSGLTVVKTFGHQTAAREQFRNVNNDVYQASFSAQ
MAV_1420|M.avium_104 QWTSVGRLNAHIEETYSGFTVVQTFGHRAAARDRFRALNDEVYRASFGAQ
MAB_1414c|M.abscessus_ATCC_199 QWANTGKLNAHIEETYSGFTIVKTYGHRAEAEAIFADRNAEVYRSAFGAQ
:* ..*:* **:******:*:*:*:**: * * * :. ::.*.**
Mflv_2250|M.gilvum_PYR-GCK LLSGLVSPATIFIGNLSYVAVAVVGGLQVATGQITLGGIQAFIQYVRQFN
Mvan_4441|M.vanbaalenii_PYR-1 FLSGLVSPATIFIGNLSYAAVAVVGGLQVATGQITLGSIQAFIQYVRQFN
MSMEG_5009|M.smegmatis_MC2_155 FFSGLVSPATMFIGNLSYVAVAVVGGLQVATGQITLGSIQAFIQYVRQFN
TH_3535|M.thermoresistible__bu FFSGLVGPVTGFIGNLSFVAVAVVGGLQVATGQVTLGSIQAFIQYVRQFN
MUL_4011|M.ulcerans_Agy99 LLSALVQPATTFIGNLGYVAVAVMGGLQVATGQITLGSVQAFIQYVRQFN
MMAR_4148|M.marinum_M LLSGLVQPATTFIGNLGYVAVAVMGGLQVATGQITLGSVQAFIQYVRQFN
Mb1303c|M.bovis_AF2122/97 FLSGLVQPATAFIGNLGYVAVAVAGGLQVATGQITLGSIQAFIQYIRQFN
Rv1272c|M.tuberculosis_H37Rv FLSGLVQPATAFIGNLGYVAVAVAGGLQVATGQITLGSIQAFIQYIRQFN
MLBr_01113|M.leprae_Br4923 FFSGLVSPATMFISNLGYVVVAALGGLQVATGHITLGNIQAFIQYVRQFN
MAV_1420|M.avium_104 FFSGLVAPATAVIGNLGYAAVAVLGGLQVATGQITLGNIQAFIQYVRQFN
MAB_1414c|M.abscessus_ATCC_199 FFSGLVSPATAFVGNLSYVAVAVVGGLKVASGGLTLGSIQAFSQYVRQFN
::*.** *.* .:.**.:..**. ***:**:* :***.:*** **:****
Mflv_2250|M.gilvum_PYR-GCK QPLTQIAGMYNTMQSGIASAERVFDLLDAEEEPADP----------VEAL
Mvan_4441|M.vanbaalenii_PYR-1 QPLTQIAGMYNTMQSGIASAERVFDLLDAEEEPSEPD--RRLAPDSPTVL
MSMEG_5009|M.smegmatis_MC2_155 QPLTQVAGMYNTLQSGIASAERVFDLLDTEEESADS--------------
TH_3535|M.thermoresistible__bu QPLTQVAGMYNTLQSGVASAERVFELLDADEESAEP--------------
MUL_4011|M.ulcerans_Agy99 TPVSQVAAMYNSLQSGVASAERVFELLDEPEEAADP----ASALQPPVSR
MMAR_4148|M.marinum_M TPVSQVAAMYNSLQSGVASAERVFELLDEPEEAADP----ASALQPPVSR
Mb1303c|M.bovis_AF2122/97 MPLSQLAGMYNALQSGVASAERVFDVLDEPEESPEP----EPELPN----
Rv1272c|M.tuberculosis_H37Rv MPLSQLAGMYNALQSGVASAERVFDVLDEPEESPEP----EPELPN----
MLBr_01113|M.leprae_Br4923 MPLSQVAGIYNTLQSGLASAERVFDLLDEPEELPDP----MLALPPP---
MAV_1420|M.avium_104 SPLSQLAGMYNTLQSGVASAERVFALLDEPEEPPDAPPEEKPTAPGP---
MAB_1414c|M.abscessus_ATCC_199 QPLTQVAAMYNTLQSGLASAERVFELLDADEETPDP------------LA
*::*:*.:**::***:******* :** ** .:.
Mflv_2250|M.gilvum_PYR-GCK PASADRRDGRVEFDGVWFSYRPGTPVVEDLSLVAEPGQTIAIVGPTGAGK
Mvan_4441|M.vanbaalenii_PYR-1 PARDGRPGGRVEFDRVWFGYEHGAPVIEDLSLVAEPGTTVAIVGPTGAGK
MSMEG_5009|M.smegmatis_MC2_155 PRRADVRTGRVEFEHVSFSYVPGTPVIEDLSLVAEPGSTVAIVGPTGAGK
TH_3535|M.thermoresistible__bu DVELPAGPGRVEFDRVSFGYRPDRPVIHDLSLVAEPGSTVAIVGPTGAGK
MUL_4011|M.ulcerans_Agy99 TGRAPALTGRVEFERVNFAYHPGHPVIHDLSLVAEPGSTVAIVGPTGAGK
MMAR_4148|M.marinum_M TGRAPALTGRVEFERVNFAYHPGHPVIHDLSLVAEPGSTVAIVGPTGAGK
Mb1303c|M.bovis_AF2122/97 ------LTGRVEFEHVNFAYLPGTPVIRDLSLVAEPGSTVAIVGPTGAGK
Rv1272c|M.tuberculosis_H37Rv ------LTGRVEFEHVNFAYLPGTPVIRDLSLVAEPGSTVAIVGPTGAGK
MLBr_01113|M.leprae_Br4923 HGQVPRINGRVEFQQVSFAYRTGTPVIDDLSMVVEPGSTVAIVGPTGAGK
MAV_1420|M.avium_104 AGRDAGGGGRVEFAHVSFAYRPGIRVIDDLSLVAEPGSTVAIVGPTGAGK
MAB_1414c|M.abscessus_ATCC_199 AATAPTGSVRVEFEDISFGYSPGQPVIEGLSLRVEPGQTVAIVGPTGAGK
**** : *.* . *: .**: .*** *:**********
Mflv_2250|M.gilvum_PYR-GCK TTLVNLLMRFYDVDSGRILLDGTDITTVSRGSLRSRIGMVLQDTWLFAGT
Mvan_4441|M.vanbaalenii_PYR-1 TTLVNLLMRFYDVDSGRILLDGVDISAVSRASLRSRIGMVLQDTWLFAGT
MSMEG_5009|M.smegmatis_MC2_155 TTLVNLLMRFYDVDSGRITIDGVDIASVSRESLRASIGMVLQDTWLFAGT
TH_3535|M.thermoresistible__bu TTLVNLLMRFYEVDSGRILIDGTDISTVSRRSLRSRIGMVLQDTWLFGGT
MUL_4011|M.ulcerans_Agy99 TTLVNLLMRFYEVDSGRILVDGIDITTVSRQSLRSHIGMVLQDTWLFDGT
MMAR_4148|M.marinum_M TTLVNLLMRFYEVDSGRILVDGIDITTVSRRSLRSHIGMVLQDTWLFDGT
Mb1303c|M.bovis_AF2122/97 TTLVNLLMRFYEIGSGRILIDGVDIASVSRQSLRSRIGMVLQDTWLYDGT
Rv1272c|M.tuberculosis_H37Rv TTLVNLLMRFYEIGSGRILIDGVDIASVSRQSLRSRIGMVLQDTWLYDGT
MLBr_01113|M.leprae_Br4923 TTLVNLLMRFYDVDSGQILIDGVDITTVSRHALRSRIGMVLQDAWLFDGT
MAV_1420|M.avium_104 TTLVNLLMRFYDVDAGRISIDGVDIATMDRRALRSRIGMVLQDTWLFDGT
MAB_1414c|M.abscessus_ATCC_199 TTLVNLLMRFYEVDSGRILLDGIDVSTMTRHDLRSRMGMVLQDTWLFGGT
***********::.:*:* :** *:::: * **: :******:**: **
Mflv_2250|M.gilvum_PYR-GCK IYDNIAYGRPDAAHDEVIEAATAAYVDRFVHTLPDGYDTRINDGGTNISA
Mvan_4441|M.vanbaalenii_PYR-1 VYENIAYGRPEAGRDEVIDAAKAAYVDRFVHTLPDGYETRIDDGGTNISA
MSMEG_5009|M.smegmatis_MC2_155 IYDNIAYGRPDADEDEVIEAATAAYVDRFVHTLPNGYDTRVDDDGGAISA
TH_3535|M.thermoresistible__bu IYDNIAYGRPDATEEEVLAAARAAYVDRFVHTLPDGYATRISDDGGAISA
MUL_4011|M.ulcerans_Agy99 ISENIAYGRPEASPDDIIQAAKAAYVDRFVHMLPASYQTRISGDGSNISA
MMAR_4148|M.marinum_M ISENIAYGRPEASPDDIIQAAKAAYVDRFVHMLPAGYQTRISGDGSNISA
Mb1303c|M.bovis_AF2122/97 IAENIAYGRPEATTDEIVEAARAAHVDRFVNTLPAGYQTRVSGDGGSISV
Rv1272c|M.tuberculosis_H37Rv IAENIAYGRPEATTDEIVEAARAAHVDRFVNTLPAGYQTRVSGDGGSISV
MLBr_01113|M.leprae_Br4923 IAENIAYGRPQASEDEVMEAAKAAYVDRFVHTLSAGYQTRVSGDGDNISA
MAV_1420|M.avium_104 IAENIGYGRPGAGDDEIVAAARAAYVDRFVQNLPDGYHTRVSGDGGNLSA
MAB_1414c|M.abscessus_ATCC_199 IVENIAYGRPGASQEEILEAARAAYVDRFVRTLPDGYDTKIADDGGNISA
: :**.**** * :::: ** **:*****. *. .* *:: ..* :*.
Mflv_2250|M.gilvum_PYR-GCK GEKQLITIARAILARPQVLVLDEATSSVDTRTEVLIQHAMRELRRDRTSF
Mvan_4441|M.vanbaalenii_PYR-1 GEKQLITIARAVLAQPQLLILDEATSSVDTRTEVLIQHAMGRLRRERTSF
MSMEG_5009|M.smegmatis_MC2_155 GEKQLITIARAVLARPKLLVLDEATSSVDTRTELLIAHAMAELRRDRTSF
TH_3535|M.thermoresistible__bu GEKQLITIARAFLARPQLLILDEATSSVDTRTEVLIQHAMTELRRDRTSF
MUL_4011|M.ulcerans_Agy99 GEKQLITIARAFLARPQLLILDEATSSVDTRTELLIARATAELRRDRTSF
MMAR_4148|M.marinum_M GEKQLITIARAFLARPQLLILDEATSSVDTRTELLIARATAELRRDRTSF
Mb1303c|M.bovis_AF2122/97 GEKQLITIARAFLARPQLLILDEATSSVDTRTELLIQRAMRELRRDRTSF
Rv1272c|M.tuberculosis_H37Rv GEKQLITIARAFLARPQLLILDEATSSVDTRTELLIQRAMRELRRDRTSF
MLBr_01113|M.leprae_Br4923 GEKQLITIARAFLARPQLLILDEATSSIDTRTEALIARATHDLLQDRTSF
MAV_1420|M.avium_104 GERQLITIARAFLARPQLLILDEATSSVDTRTEALIAHATRELRRDRTSF
MAB_1414c|M.abscessus_ATCC_199 GEKQLITIARAFLARPQLLILDEATSSVDTRTELRIQRAMAELRRDRTSF
**:********.**:*::*:*******:***** * :* * ::****
Mflv_2250|M.gilvum_PYR-GCK IIAHRLSTIRDADTILVMEAGRIVERGTHSELLARRGEYWTMVNR
Mvan_4441|M.vanbaalenii_PYR-1 IIAHRLSTIRDADLILVMQAGRIVERGTHEELLARGGEYAAMVNR
MSMEG_5009|M.smegmatis_MC2_155 IIAHRLSTIRDADLILVMDSGRIIERGTHEELLARHGRYWEMTRV
TH_3535|M.thermoresistible__bu IIAHRLSTVRDADLILVLEEGRIVERGSHDELMARRGEYWAMTQA
MUL_4011|M.ulcerans_Agy99 IIAHRLSTIRDADQILVVQAGQIVEHGTHAELLANRGAYYAMTRA
MMAR_4148|M.marinum_M IIAHRLSTIRDADQILVVQAGQIVEHGTHAELLANRGAYYAMTRA
Mb1303c|M.bovis_AF2122/97 IIAHRLSTIRDADHILVVQTGQIVERGNHAELLARRGVYYQMTRA
Rv1272c|M.tuberculosis_H37Rv IIAHRLSTIRDADHILVVQTGQIVERGNHAELLARRGVYYQMTRA
MLBr_01113|M.leprae_Br4923 IIAHHLPTIRDADHILVVAAGKIVEQGSHAKLLARRGAYYRMTQA
MAV_1420|M.avium_104 IIAHRLSTIRDADRIAVIEAGRIFEQGTHAELLARRGVYYAMTQA
MAB_1414c|M.abscessus_ATCC_199 IIAHRLSTIRDADRIVVLDGGRVTESGTHTELLARHGAYFAMTQS
****:*.*:**** * *: *:: * *.* :*:*. * * *..