For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LDPLVPGAPVLVAGARVTGRAVLAALTRFGAIPTMCDDDPAMLRPFAESGVATVDPSVAVQQLAQPEPSS ESRYALVVTSPGFQPSTPVLAAAAAAGVPIWGDVELAWRLDAAGCYGPPRRWLVVTGTNGKTTTTSMLHA MLVAGQHRSVLCGNIGSPVLDVLEEPADLLAVELSSFQLRWAPSLRPEAGVVLNIAEDHLDWHGTMAAYT EAKARVLTGRVAVVGLDDSRAAALRDTAPAPVRVGFRIGTPAVGELGLRDGHLVDRAFSDDLVLLPASSI PVPGPVGVLDTLAAAALARAVGVSADAIADAVASFQVGRHRSEVVEVVDGITYVDDSKATNPHAAEASVL AYPRVVWVAGGLLKGASVDAEVARMAARLVGAVLIGQDRAEVAEALSRHAPDVPVVQVVTSEDADMGEAY GASVTKVDDVGGSLGARVMRAAVAAARNMARSGDTVLLAPAGASFDQFTSYGDRGDAFAAAVRAAVR*
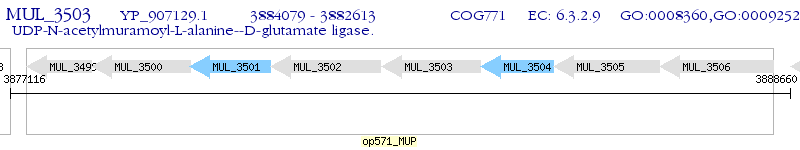
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3503 | murD | - | 100% (488) | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase |
| M. ulcerans Agy99 | MUL_3506 | murE | 8e-20 | 29.84% (429) | UDP-N-acetylmuramoylalanyl-D-glutamate--2,6-diaminopimelate |
| M. ulcerans Agy99 | MUL_3505 | murF | 3e-15 | 26.21% (515) | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6- |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2179c | murD | 0.0 | 77.94% (494) | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase |
| M. gilvum PYR-GCK | Mflv_2986 | murD | 1e-180 | 68.25% (485) | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase |
| M. tuberculosis H37Rv | Rv2155c | murD | 0.0 | 77.73% (494) | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase |
| M. leprae Br4923 | MLBr_00912 | murD | 0.0 | 76.52% (494) | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase |
| M. abscessus ATCC 19977 | MAB_2004 | murD | 1e-158 | 61.65% (485) | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase |
| M. marinum M | MMAR_3195 | murD | 0.0 | 98.77% (488) | UDP-N-acetylmuramoylalanine-D-glutamate ligase MurD |
| M. avium 104 | MAV_2334 | murD | 0.0 | 77.55% (490) | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase |
| M. smegmatis MC2 155 | MSMEG_4229 | murD | 0.0 | 68.97% (493) | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase |
| M. thermoresistible (build 8) | TH_1516 | - | 1e-73 | 72.83% (184) | Probable UDP-N-acetylmuramoylalanine-D-glutamate ligase MurD |
| M. vanbaalenii PYR-1 | Mvan_3525 | murD | 1e-178 | 67.01% (491) | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2986|M.gilvum_PYR-GCK ---MSGPDVDDLAGLSVLVTGARVTGQAILAALT---PLGARATLTDDSP
Mvan_3525|M.vanbaalenii_PYR-1 ---MQP----ISPGAAVLVVGAGITGRAVLAALA---PLGARATLTDDSP
MSMEG_4229|M.smegmatis_MC2_155 MPDVTGDLAPLTPGARVLVTGAGITGRAVLGALA---PLEVAATLCDDNA
MUL_3503|M.ulcerans_Agy99 ------MLDPLVPGAPVLVAGARVTGRAVLAALT---RFGAIPTMCDDDP
MMAR_3195|M.marinum_M ------MLDPLVPGAPVLVAGARVTGRAVLAALT---RFGAIPTMCDDDP
Mb2179c|M.bovis_AF2122/97 ------MLDPLGPGAPVLVAGGRVTGQAVAAVLT---RFGATPTVCDDDP
Rv2155c|M.tuberculosis_H37Rv ------MLDPLGPGAPVLVAGGRVTGQAVAAVLT---RFGATPTVCDDDP
MLBr_00912|M.leprae_Br4923 ------MLDTLVPGAPVLVAGGGVTGRAVLAALT---RFGMAATLCDDDP
MAV_2334|M.avium_104 MSAAEPALAPLSPGAPVLVAGGGITGRAVLAALR---RFGAAPTLCDDDP
TH_1516|M.thermoresistible__bu --------------------------------------------------
MAB_2004|M.abscessus_ATCC_1997 --MTTCEVAELTGGTRVLVCGARVTGDAVVAALTGLADLGLRITVCDDDE
Mflv_2986|M.gilvum_PYR-GCK TALTAFAQQGVEVLDPAAAVERIAA--------FDLVVTSPGFPPTAPVL
Mvan_3525|M.vanbaalenii_PYR-1 TALTASAQQRVEVLDPATAVDRIAD--------FDLVVVSPGIPPTAPIP
MSMEG_4229|M.smegmatis_MC2_155 DALAALAAKGVAVIAPGDAIASIGD--------YALVVTSPGFAPSAPVL
MUL_3503|M.ulcerans_Agy99 AMLRPFAESGVATVDPSVAVQQLAQPEPSSESRYALVVTSPGFQPSTPVL
MMAR_3195|M.marinum_M AMLRPFAESGVATVDPSVAVQQLAQPEASPESRYALVVTSPGFQPSTPVL
Mb2179c|M.bovis_AF2122/97 VMLRPHAERGLPTVSSSDAVQQITG--------YALVVASPGFSPATPLL
Rv2155c|M.tuberculosis_H37Rv VMLRPHAERGLPTVSSSDAVQQITG--------YALVVASPGFSPATPLL
MLBr_00912|M.leprae_Br4923 AALQQYADNGVATVSAATATQQMFE----RGRKYVLVVTSPGFAPTTPVL
MAV_2334|M.avium_104 ATLRGYVDSGVDTVSTSAAAERISR--------YALVVTSPGFAPTAPLP
TH_1516|M.thermoresistible__bu --------------------------------------------------
MAB_2004|M.abscessus_ATCC_1997 DALQRHRDAGHGALTTAAAQRHMAD--------QDLVIVSPGFGPSAPVV
Mflv_2986|M.gilvum_PYR-GCK AAAADAGVPIWGDVELAWRLDRSGRFGPPRQWLVVTGTNGKTTTTSMLHA
Mvan_3525|M.vanbaalenii_PYR-1 VAAAAAGIPVWGDVELAWHLDRSGRFGPPRQWLVVTGTNGKTTTTSMLYA
MSMEG_4229|M.smegmatis_MC2_155 AAAAAAGVPIWGDVELAWRLDAAGRYGPPRRWLVVTGTNGKTTTTSMLHD
MUL_3503|M.ulcerans_Agy99 AAAAAAGVPIWGDVELAWRLDAAGCYGPPRRWLVVTGTNGKTTTTSMLHA
MMAR_3195|M.marinum_M AAAAAAGVPIWGDVELAWRLDAAGCYGPPRRWLVVTGTNGKTTTTSMLHA
Mb2179c|M.bovis_AF2122/97 AAAAAAGVPIWGDVELAWRLDAAGCYGPPRSWLVVTGTNGKTTTTSMLHA
Rv2155c|M.tuberculosis_H37Rv AAAAAAGVPIWGDVELAWRLDAAGCYGPPRSWLVVTGTNGKTTTTSMLHA
MLBr_00912|M.leprae_Br4923 VAASAARVPIWGDVELAWRLDAAGYYGPPRRWLVVTGTNGKTTTTSMLHA
MAV_2334|M.avium_104 LAAAAAGVPVWGDVELAWRLDAAGHYGPPRRWLVVTGTNGKTTTTSMLHA
TH_1516|M.thermoresistible__bu -------XPVWGDVELAWRLDAAGRFGAPRRWLVVTGTNGKTTTTSMLQA
MAB_2004|M.abscessus_ATCC_1997 QAAVQAGLPIWGDVELAWRLDASGRYGRPKRWVVVTGTNGKTTTTAMVHD
*:********:** :* :* *: *:************:*:
Mflv_2986|M.gilvum_PYR-GCK MLESAGRRAVLCGNIGDPVLTLL---EQPADVFAVELSSFQLHWAPSLRP
Mvan_3525|M.vanbaalenii_PYR-1 MLQAGGRRAVLCGNIGDPVLSQL---DRPADVLAVELSSFQLHWAPSLRP
MSMEG_4229|M.smegmatis_MC2_155 MLVAGGRRSVLCGNIGDPVLAVL---DQPAELLAVELSSFQLHWAPSLRP
MUL_3503|M.ulcerans_Agy99 MLVAGQHRSVLCGNIGSPVLDVL---EEPADLLAVELSSFQLRWAPSLRP
MMAR_3195|M.marinum_M MLVAGQHRSVLCGNIGSPVLDVL---EEPADLLAVELSSFQLHWAPSLRP
Mb2179c|M.bovis_AF2122/97 MLIAGGRRAVLCGNIGSAVLDVL---DEPAELLAVELSSFQLHWAPSLRP
Rv2155c|M.tuberculosis_H37Rv MLIAGGRRAVLCGNIGSAVLDVL---DEPAELLAVELSSFQLHWAPSLRP
MLBr_00912|M.leprae_Br4923 MLAADNRRSLLCGNIGRPVLDVLTEFAEPSDFLAVELSSFQLHWAPSLRP
MAV_2334|M.avium_104 MLTAAGRRSLLCGNIGSPVLDVL---DQPAELLAVELSSFQLHWAPSLRP
TH_1516|M.thermoresistible__bu MLLADGRRSALCGNIGNPVLDVL---TEPVDLFAVELSSFQLHWAPSVRP
MAB_2004|M.abscessus_ATCC_1997 MLTAAGGTSALCGNIGDPVLNVL---DRDVEFLAVELSSFQLHWAPSVRP
** : : ****** .** * . :.:*********:****:**
Mflv_2986|M.gilvum_PYR-GCK EAGAVLNVAEDHLDWHGSMAAYAADKARALTGRVAVGGLDDEIAAGLLGS
Mvan_3525|M.vanbaalenii_PYR-1 EAGAVLNVAEDHLDWHGSMAAYAGDKARALTGRVAVGGLDDAIAAGLLAD
MSMEG_4229|M.smegmatis_MC2_155 EAGVVLNVAEDHLDWHGSMAAYARDKARVLDGRVAVVGLDDPVAAGLLPT
MUL_3503|M.ulcerans_Agy99 EAGVVLNIAEDHLDWHGTMAAYTEAKARVLTGRVAVVGLDDSRAAALRDT
MMAR_3195|M.marinum_M EAGVVLNIAEDHLDWHGTMAAYTEAKARVLTGRVAVVGLDDSRAAALRDT
Mb2179c|M.bovis_AF2122/97 EAGAVLNIAEDHLDWHATMAEYTAAKARVLTGGVAVAGLDDSRAAALLDG
Rv2155c|M.tuberculosis_H37Rv EAGAVLNIAEDHLDWHATMAEYTAAKARVLTGGVAVAGLDDSRAAALLDG
MLBr_00912|M.leprae_Br4923 EAGVVLNIAEDHLDWHSTMADYTMAKARVLTGRVAVVGLDDSRAAALLST
MAV_2334|M.avium_104 EGGAVLNIAEDHLDWHGTLADYAAAKARVLDGRVAVVGLDDSRAAALLST
TH_1516|M.thermoresistible__bu YAGVVLNIAEDHLDWHGSLTAYAQAKARVLGGTVAVAGVDDPMAARLLAA
MAB_2004|M.abscessus_ATCC_1997 VAGAVLNVAEDHLDWHGGMEGYTRDKARALDGEVAVAGLDDAIAGGLLAA
.*.***:********. : *: ***.* * *** *:** *. *
Mflv_2986|M.gilvum_PYR-GCK APAAVKVGFRLGAPGPGEFGVRDGALIDNAFADGLELAAVDSIPVAGPVG
Mvan_3525|M.vanbaalenii_PYR-1 APAPVKVGFRLGDPGPGEFGVRDGVLIDNAFGRDLALADADTIPVAGPVG
MSMEG_4229|M.smegmatis_MC2_155 AAATVRVGFRLGDPAAGELGVRDGKLIDRAFDDGVELADCADIGVAGPIG
MUL_3503|M.ulcerans_Agy99 APAPVRVGFRIGTPAVGELGLRDGHLVDRAFSDDLVLLPASSIPVPGPVG
MMAR_3195|M.marinum_M APAPVRVGFRIGAPAVGELGLRDGHLVDRAFSDDLVLLPASSIPVPGPVG
Mb2179c|M.bovis_AF2122/97 SPAQVRVGFRLGEPAAGELGVRDAHLVDRAFSDDLTLLPVASIPVPGPVG
Rv2155c|M.tuberculosis_H37Rv SPAQVRVGFRLGEPAARELGVRDAHLVDRAFSDDLTLLPVASIPVPGPVG
MLBr_00912|M.leprae_Br4923 TVAPVRVGFRFGEPAVGELGVRDGYLVDRAFAEDLALMPVTSIPVSGPVG
MAV_2334|M.avium_104 ARAPVRVGFRLGEPAAGELGVRGGQLVDRAFADDLTLLPVDSIPVPGPVG
TH_1516|M.thermoresistible__bu AAAPRKVGFRLGAPAADELGVADGMLVDRAFADGLPLVAVDRIPVAXXX-
MAB_2004|M.abscessus_ATCC_1997 AAAPVKAGFRAGEPAVGELGVLDGWLVDRAFGVDIPLIEVSKLPVGGPIG
: * :.*** * *. *:*: .. *:*.** .: * : *
Mflv_2986|M.gilvum_PYR-GCK VLNALAAAALARAVDVPAEAIATALAGFQVGRHRAEVVAVADGITYVDDS
Mvan_3525|M.vanbaalenii_PYR-1 VLDALAAAALARAVDVPAAAIAEALATFHVGRHRAEVVAVADGITYVDDS
MSMEG_4229|M.smegmatis_MC2_155 VLDALGAAALARAVGVAPGAIGAALASFQVGRHRAEVVGTVDGVVFIDDS
MUL_3503|M.ulcerans_Agy99 VLDTLAAAALARAVGVSADAIADAVASFQVGRHRSEVVEVVDGITYVDDS
MMAR_3195|M.marinum_M VLDTLAAAALARAVGVSADAIADAVASFQVGRHRSEVVEVVDGITYVDDS
Mb2179c|M.bovis_AF2122/97 VLDALAAAALARSVGVPAGAIADAVTSFRVGRHRAEVVAVADGITYVDDS
Rv2155c|M.tuberculosis_H37Rv VLDALAAAALARSVGVPAGAIADAVTSFRVGRHRAEVVAVADGITYVDDS
MLBr_00912|M.leprae_Br4923 VLDALAAAALARSVGVGATAIADAVALFRLGRHRAEVVAVADGIRYVDDS
MAV_2334|M.avium_104 VLDALAAAALARCVDVPASPIAEAIVSFRVGRHRAEVVAVADGITYVDDS
TH_1516|M.thermoresistible__bu -------------------------------------XRVLHG-------
MAB_2004|M.abscessus_ATCC_1997 VLNALAAAALARAVGTPAVHIARALEVFQVGRHRAELIARVADVAYVDDS
.
Mflv_2986|M.gilvum_PYR-GCK KATNPHAARASIDAYPRVVWIAGGMLKGASVEDLVSDSADHLVGAVLIGR
Mvan_3525|M.vanbaalenii_PYR-1 KATNPHAAQASVSAFPRVVWVAGGLLKGASVDEMVEATKDRLVGVVLIGR
MSMEG_4229|M.smegmatis_MC2_155 KATNPHAAQASITAYDRVVWVAGGLLKGASVDELVRQVANRLAGAVLIGR
MUL_3503|M.ulcerans_Agy99 KATNPHAAEASVLAYPRVVWVAGGLLKGASVDAEVARMAARLVGAVLIGQ
MMAR_3195|M.marinum_M KATNPHAAEASVLAYPRVVWVAGGLLKGASVDAEVARMASRLVGAVLIGQ
Mb2179c|M.bovis_AF2122/97 KATNPHAARASVLAYPRVVWIAGGLLKGASLHAEVAAMASRLVGAVLIGR
Rv2155c|M.tuberculosis_H37Rv KATNPHAARASVLAYPRVVWIAGGLLKGASLHAEVAAMASRLVGAVLIGR
MLBr_00912|M.leprae_Br4923 KATNPHAALVSVLAYPRVVWVAGGLLKGASVDAEVARMAPQLVGAVLIGR
MAV_2334|M.avium_104 KATNPHAAEASVLAYPRVVWIAGGLLKGASVDAVVARMASRLVGAVLIGR
TH_1516|M.thermoresistible__bu -EVDPHPARR----------------------------------------
MAB_2004|M.abscessus_ATCC_1997 KATNPHAAAASIAGYPRVVWIAGGLLKGAAVDDLVRDCADRLGAVVLIGR
.:**.*
Mflv_2986|M.gilvum_PYR-GCK DRAVVAEALARHAPDVPVVELVTGEDSGVLEKDESGVTR---VTRVVQAD
Mvan_3525|M.vanbaalenii_PYR-1 DRAIVGNALSRHAPDVPVVELVTGEDSVVLEKDESGVTR---VTRVIQTG
MSMEG_4229|M.smegmatis_MC2_155 DRQMVADALSRHAPDVPVVELVAGEDSGVLGTNESIESVGDHVTRVIEVG
MUL_3503|M.ulcerans_Agy99 DRAEVAEALSRHAPDVPVVQVVTSEDADMG------EAYGASVTKVDDVG
MMAR_3195|M.marinum_M DRAEVAEALSRHAPDVPVVQVVTSEDADMG------EACGASVTKVDDVG
Mb2179c|M.bovis_AF2122/97 DRAAVAEALSRHAPDVPVVQVVAGEDTGMPATVEVPVACVLDVAKDDKAG
Rv2155c|M.tuberculosis_H37Rv DRAAVAEALSRHAPDVPVVQVVAGEDTGMPATVEVPVACVLDVAKDDKAG
MLBr_00912|M.leprae_Br4923 DRAMVAEALSRHAPNVPVVQVVAGEDAGMHA---VAVVSGTDVISISDVG
MAV_2334|M.avium_104 DRREVAEALSRHAPDVPVVHVVTGEDAGMDAT---PVVFGANVTKVKHLG
TH_1516|M.thermoresistible__bu --------------------------------------------------
MAB_2004|M.abscessus_ATCC_1997 DRALIAEALARHAPDVPVVAVVTREDAGVQ------EANVVYVTDEADGG
Mflv_2986|M.gilvum_PYR-GCK SESLSDAVMAAAVRAARGLAAPGDAILLAPAGASFDQFTSYGHRGDTFAA
Mvan_3525|M.vanbaalenii_PYR-1 DRTLADAVMTAAVDAARGLATSGDTVLLAPAGASFDQFTGYGHRGDAFAA
MSMEG_4229|M.smegmatis_MC2_155 GRPVSDAVMSAVVSAARDLASPGDTVLLAPAGASFDQFSGYGQRGDAFAA
MUL_3503|M.ulcerans_Agy99 -GSLGARVMRAAVAAARNMARSGDTVLLAPAGASFDQFTSYGDRGDAFAA
MMAR_3195|M.marinum_M -GSLGARVMRAAVAAARNMARSGDTVLLAPAGASFDQFTSYGDRGDAFAA
Mb2179c|M.bovis_AF2122/97 -ETVGAAVMTAAVAAARRMAQPGDTVLLAPAGASFDQFTGYADRGEAFAT
Rv2155c|M.tuberculosis_H37Rv -ETVGAAVMTAAVAAARRMAQPGDTVLLAPAGASFDQFTGYADRGEAFAT
MLBr_00912|M.leprae_Br4923 -GTIGTRVMVAAVAAARDLAQPGDTVLLAPAGASFDQFSGYADRGDTFAT
MAV_2334|M.avium_104 -GDLGAAVMSAAVAAARDLAKPGDTVLLAPAAASFDQFAGYADRGDAFAA
TH_1516|M.thermoresistible__bu --------------------------------------------------
MAB_2004|M.abscessus_ATCC_1997 GADLGTRVMAAAVRHASSLARPGDTVLLAPAGASFDQFSGYSERGDRFAA
Mflv_2986|M.gilvum_PYR-GCK AVRAALR
Mvan_3525|M.vanbaalenii_PYR-1 AVRAAIR
MSMEG_4229|M.smegmatis_MC2_155 AVRAAVG
MUL_3503|M.ulcerans_Agy99 AVRAAVR
MMAR_3195|M.marinum_M AVRAAVR
Mb2179c|M.bovis_AF2122/97 AVRAVIR
Rv2155c|M.tuberculosis_H37Rv AVRAVIR
MLBr_00912|M.leprae_Br4923 AVRAAIR
MAV_2334|M.avium_104 AVRAAIR
TH_1516|M.thermoresistible__bu -------
MAB_2004|M.abscessus_ATCC_1997 AVHALTG