For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSIENLKAALPEYAKDLKLNLGSISRTTVLDEEQLWGTLLASAAATRNAQVLAEIGAEAADNLSAQAYQA ALGAVSIMGMNNVFYRGRGFLEGQYDDLRAGLRMNIIANPGVDKANFELWSFAVSSVNGCSHCVVAHEHT LREAGVGREAVFEALKAAAIVCGVAQALTAAQTLAAVG
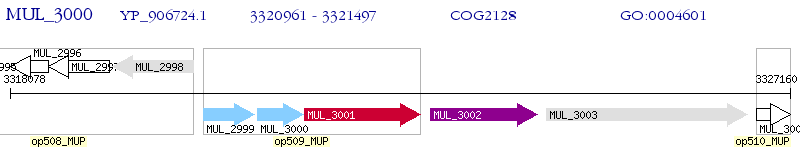
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3000 | ahpD | - | 100% (178) | alkyl hydroperoxide reductase D protein AhpD |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2455 | ahpD | 6e-78 | 80.57% (175) | alkyl hydroperoxide reductase subunit D |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv2429 | ahpD | 6e-78 | 80.57% (175) | alkyl hydroperoxide reductase subunit D |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4407c | - | 2e-65 | 70.69% (174) | putative alkylhydroperoxidase AhpD |
| M. marinum M | MMAR_2754 | ahpD | 6e-96 | 99.44% (178) | alkyl hydroperoxide reductase D protein AhpD |
| M. avium 104 | MAV_2840 | ahpD | 3e-79 | 79.78% (178) | alkylhydroperoxidase, AhpD family protein |
| M. smegmatis MC2 155 | MSMEG_4890 | ahpD | 6e-67 | 68.93% (177) | alkylhydroperoxidase, AhpD family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_3000|M.ulcerans_Agy99 MSIENLKAALPEYAKDLKLNLGSISRTTVLDEEQLWGTLLASAAATRNAQ
MMAR_2754|M.marinum_M MSIENLKAALPEYAKDLKLNLGSISRTTVLDEEQLWGTLLASAAATRNAQ
Mb2455|M.bovis_AF2122/97 MSIEKLKAALPEYAKDIKLNLSSITRSSVLDQEQLWGTLLASAAATRNPQ
Rv2429|M.tuberculosis_H37Rv MSIEKLKAALPEYAKDIKLNLSSITRSSVLDQEQLWGTLLASAAATRNPQ
MAV_2840|M.avium_104 MSVENLKEALPEYAKDLKLNLGSITRTTELNEEQLWGTLLASAAATRNTQ
MAB_4407c|M.abscessus_ATCC_199 MSIENLKEALPEYAKDLKLNLGTVARGTVLSQSQLWGTLVATAAATRNEQ
MSMEG_4890|M.smegmatis_MC2_155 MSIDNIKSALPEYAKDLKLNLGSIANTTELTEQQLWGALVATAAATKNAR
**::::* ********:****.:::. : * :.****:*:*:****:* :
MUL_3000|M.ulcerans_Agy99 VLAEIGAEAADNLSAQAYQAALGAVSIMGMNNVFYRGRGFLEGQYDDLRA
MMAR_2754|M.marinum_M VLAEIGAEAADNLSAQAYQAALGAVSIMGMNNVFYRGRGFLEGQYDDLRA
Mb2455|M.bovis_AF2122/97 VLADIGAEATDHLSAAARHAALGAAAIMGMNNVFYRGRGFLEGRYDDLRP
Rv2429|M.tuberculosis_H37Rv VLADIGAEATDHLSAAARHAALGAAAIMGMNNVFYRGRGFLEGRYDDLRP
MAV_2840|M.avium_104 VLTEIGAEAADTLSAEAYHAALGAASVMAMNNVFYRGRGFLDGKYDDLRA
MAB_4407c|M.abscessus_ATCC_199 VFKEIREEAAGILTPEALDAALGAASIMAMTNVFYRGRGFLDGAYDDLRP
MSMEG_4890|M.smegmatis_MC2_155 LLREISEDALDILSEEAYNAALGAAAIMGMNNVFYRTKGQLDGKYDDLRA
:: :* :* . *: * .*****.::*.*.***** :* *:* *****.
MUL_3000|M.ulcerans_Agy99 GLRMNIIANPGVDKANFELWSFAVSSVNGCSHCVVAHEHTLREAGVGREA
MMAR_2754|M.marinum_M GLRMNIIANPGVDKANFELWSFAVSSVNGCSHCVVAHEHTLREAGVGREA
Mb2455|M.bovis_AF2122/97 GLRMNIIANPGIPKANFELWSFAVSAINGCSHCLVAHEHTLRTVGVDREA
Rv2429|M.tuberculosis_H37Rv GLRMNIIANPGIPKANFELWSFAVSAINGCSHCLVAHEHTLRTVGVDREA
MAV_2840|M.avium_104 GLRMNIIGNPGVEKANFELWCFAVSAINGCPDCVASHEHTLREAGVSRET
MAB_4407c|M.abscessus_ATCC_199 GLRMNIIGNPGVDKSEFELWSFAVSTINGCHYCVGSHEKVLREAGLTREQ
MSMEG_4890|M.smegmatis_MC2_155 GLRMNIIGNPGVAKEDFELWSLAVSAINGCGHCLAAHEKTLRDADVARTT
*******.***: * :****.:***::*** *: :**:.** ..: *
MUL_3000|M.ulcerans_Agy99 VFEALKAAAIVCGVAQALTAAQTLAAVG
MMAR_2754|M.marinum_M VLEALKAAAIVCGVAQALTAAQTLAAVG
Mb2455|M.bovis_AF2122/97 IFEALKAAAIVSGVAQALATIEALSPS-
Rv2429|M.tuberculosis_H37Rv IFEALKAAAIVSGVAQALATIEALSPS-
MAV_2840|M.avium_104 IQEALKAAAIISGVAQAIVTSQTLATAG
MAB_4407c|M.abscessus_ATCC_199 VLEALKIAAIVAGVAQALATVPALV---
MSMEG_4890|M.smegmatis_MC2_155 IFEAIRLASIVSGVAQALLTADTLAAV-
: **:: *:*:.*****: : :*