For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AKLVVTPQYLQTLAATQTQAAATAASAGAASTNLKNEVWVSHGVISGASNTSFANAEAARRAAANSMNTT SSKLAAKLRTASSAYQGVDEQAGESLDKQIIER*
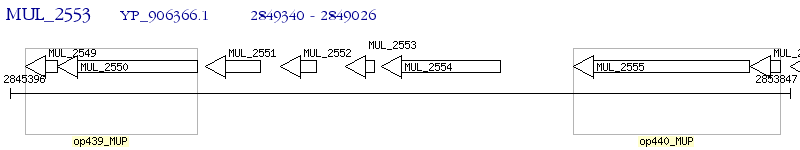
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2553 | - | - | 100% (104) | hypothetical protein MUL_2553 |
| M. ulcerans Agy99 | MUL_4591 | - | 1e-05 | 24.49% (98) | hypothetical protein MUL_4591 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3895 | - | 9e-05 | 23.96% (96) | hypothetical protein Mb3895 |
| M. gilvum PYR-GCK | Mflv_0157 | - | 2e-10 | 33.33% (90) | hypothetical protein Mflv_0157 |
| M. tuberculosis H37Rv | Rv3865 | - | 9e-05 | 23.96% (96) | hypothetical protein Rv3865 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1326 | - | 2e-51 | 99.04% (104) | hypothetical protein MMAR_1326 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0728 | - | 2e-12 | 38.04% (92) | hypothetical protein MSMEG_0728 |
| M. thermoresistible (build 8) | TH_1469 | - | 6e-07 | 30.43% (92) | CONSERVED HYPOTHETICAL PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_0655 | - | 1e-07 | 30.21% (96) | hypothetical protein Mvan_0655 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3895|M.bovis_AF2122/97 --------------------------------------------------
Rv3865|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2553|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1326|M.marinum_M --------------------------------------------------
MSMEG_0728|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0157|M.gilvum_PYR-GCK MALGKLWVDANYFYGVADHANMMSSDPALGSAAIATDAAILMQSLAAVGG
Mvan_0655|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1469|M.thermoresistible__bu --------------------------------------------------
Mb3895|M.bovis_AF2122/97 --------------------------------------------------
Rv3865|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2553|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1326|M.marinum_M --------------------------------------------------
MSMEG_0728|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0157|M.gilvum_PYR-GCK STMGATGLATSVATPIINAGLMTLTAASNTLGFGRPEDGERFTDGARQFS
Mvan_0655|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1469|M.thermoresistible__bu --------------------------------------------------
Mb3895|M.bovis_AF2122/97 --------------------------------------------------
Rv3865|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2553|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1326|M.marinum_M --------------------------------------------------
MSMEG_0728|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0157|M.gilvum_PYR-GCK EANDELQQAVPPPDWQGDASDAYGLRNTEQQQRAGSMATFDGQIQEVLAE
Mvan_0655|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1469|M.thermoresistible__bu -----------------LSGNVFVQPEGLQTFSQTHSGIAGALPQSSAEA
Mb3895|M.bovis_AF2122/97 --------------------------------------------------
Rv3865|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2553|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1326|M.marinum_M --------------------------------------------------
MSMEG_0728|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0157|M.gilvum_PYR-GCK EAGQVQDTRDFVSTRQVMLAAAIPAAITAKLIPPGGAAAAMAIELAAVAA
Mvan_0655|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1469|M.thermoresistible__bu TPGPVETSHGPIAAAANAALRNALSSRGGTVQATTTSAATISELLHRAAR
Mb3895|M.bovis_AF2122/97 ------------------------------------------------MT
Rv3865|M.tuberculosis_H37Rv ------------------------------------------------MT
MUL_2553|M.ulcerans_Agy99 -------------------------------------------------M
MMAR_1326|M.marinum_M -------------------------------------------------M
MSMEG_0728|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0157|M.gilvum_PYR-GCK TVPFAMQRVTEMVSHSADNADRIRQIGSHYGVVDESAQMPGGLSGAQSGT
Mvan_0655|M.vanbaalenii_PYR-1 ---------------------------------------------MTSPD
TH_1469|M.thermoresistible__bu AYQEGDLRSAEELRAAANAIADGTGAGGXX-XAHERVRKAAKAADRAAGD
Mb3895|M.bovis_AF2122/97 GFLGVVPSFLKVLAGMHNEIVGDIKRATDTVAGISGRVQLTHGSFTSKFN
Rv3865|M.tuberculosis_H37Rv GFLGVVPSFLKVLAGMHNEIVGDIKRATDTVAGISGRVQLTHGSFTSKFN
MUL_2553|M.ulcerans_Agy99 AKLVVTPQYLQTLAATQTQAAATAASAGAASTNLKNEVWVSHGVISGASN
MMAR_1326|M.marinum_M ANLVVTPQYLQTLAATQTQAAATAASAGAASTNLKNEVWVSHGVISGASN
MSMEG_0728|M.smegmatis_MC2_155 ----MITGYVQELAELQGRTAQQILSALEAAQGVGTSMWKNHGMVCAPSN
Mflv_0157|M.gilvum_PYR-GCK PAPLRVSESIRGTAGIQQNVAATIDSARSAPDGSAGDVFKTHGVVCTSTA
Mvan_0655|M.vanbaalenii_PYR-1 GELHVEPSHLRQLSARQNAAAAQIADAVPLTADESDHVLSSHGIVCMPTY
TH_1469|M.thermoresistible__bu DERDEALHRIRKGAKRLRYLAAAAGAAEVADRAEAVQTLLGEHQDSVVSR
:: : . * .
Mb3895|M.bovis_AF2122/97 DTLQEFETTRSSTGTGLQGVTSGLANNLLAAAGAYLKADDGLAGVIDKIF
Rv3865|M.tuberculosis_H37Rv DTLQEFETTRSSTGTGLQGVTSGLANNLLAAAGAYLKADDGLAGVIDKIF
MUL_2553|M.ulcerans_Agy99 TSFANAEAARRAAANSMNTTSSKLAAKLRTASSAYQGVDEQAGESLDKQI
MMAR_1326|M.marinum_M TSFANAEAARRAAANSMNTTSSKLAAKLRTASSAYQGVDEQAGESLDKQI
MSMEG_0728|M.smegmatis_MC2_155 SAVIAAETARRAACTRMNAKSEELAQKLGMARKLYDGTDMQEEQKLDDEM
Mflv_0157|M.gilvum_PYR-GCK AALKSAELARRSAAEKMHLISTDLSAKLGSSATEYSHTEQQNADAVDRQM
Mvan_0655|M.vanbaalenii_PYR-1 LAAQAAGVARAKAGVAVQVMSEAFAANLDSAAANYSWTDQDSGSNIDGKM
TH_1469|M.thermoresistible__bu AHLIAQADAAHAAGADTFTYGLLYQQEVALAQRCRAQLDPALKKLRKAVR
: : :: : : .
Mb3895|M.bovis_AF2122/97 G-----
Rv3865|M.tuberculosis_H37Rv G-----
MUL_2553|M.ulcerans_Agy99 IER---
MMAR_1326|M.marinum_M IER---
MSMEG_0728|M.smegmatis_MC2_155 RCTPSG
Mflv_0157|M.gilvum_PYR-GCK PPR---
Mvan_0655|M.vanbaalenii_PYR-1 RPGRR-
TH_1469|M.thermoresistible__bu RYRKRG