For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TQKNGETGTSTDSRDDVAEVVSRLIRFDTTNTGEPETTKGEAECARWVAEQLAEVGYQTEYVESGAPGRG NVFARLRGADSSRGALLIHGHLDVVPAEAADWSVHPFSGAIEDGYVWGRGAVDMKDMVGMMIVVARHFRR AGIVPPRDLVFAFLADEEHGGHFGAHWLVDNRPDLFDGVTEAVGEVGGFSLTVPRRDGGERRLYLVETAE KGILWMRLTAQGRAGHGSMVHDHNAVTTVAEAVARLGRHQFPLVITDTVAQFLHAVSEETGLDLDPDSPD LEGTLDKLGPIARMLKAVLRDTANPTMLKAGYKANVIPATAEAVVDCRVLPGRQAAFEAEVDELIGPDVS REWISELASYETSFDGDLVDAMNAAVLSVDPDGRTVPYMLSGGTDAKAFARLGIRCFGFSPLRLPPELDF ATLFHGVDERVPIDALRFGTEVLTHFLTHC*
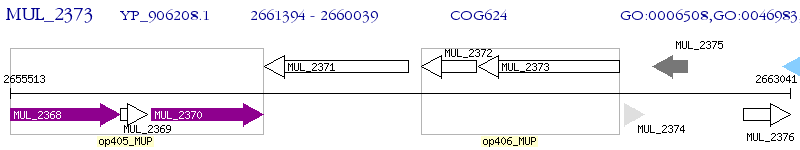
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2373 | - | - | 100% (451) | hypothetical protein MUL_2373 |
| M. ulcerans Agy99 | MUL_3812 | argE | 5e-05 | 27.62% (181) | hypothetical protein MUL_3812 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2165c | - | 0.0 | 88.09% (445) | hypothetical protein Mb2165c |
| M. gilvum PYR-GCK | Mflv_3037 | - | 0.0 | 77.98% (445) | hypothetical protein Mflv_3037 |
| M. tuberculosis H37Rv | Rv2141c | - | 0.0 | 88.09% (445) | hypothetical protein Rv2141c |
| M. leprae Br4923 | MLBr_01288 | - | 0.0 | 82.47% (445) | hypothetical protein MLBr_01288 |
| M. abscessus ATCC 19977 | MAB_2102 | - | 0.0 | 78.68% (441) | hypothetical protein MAB_2102 |
| M. marinum M | MMAR_3123 | - | 0.0 | 99.33% (451) | hypothetical protein MMAR_3123 |
| M. avium 104 | MAV_2350 | - | 0.0 | 82.93% (451) | hypothetical protein MAV_2350 |
| M. smegmatis MC2 155 | MSMEG_4200 | - | 0.0 | 78.68% (441) | hypothetical protein MSMEG_4200 |
| M. thermoresistible (build 8) | TH_2370 | dapE2 | 0.0 | 78.90% (436) | CONSERVED HYPOTHETICAL PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_3489 | - | 0.0 | 80.89% (429) | hypothetical protein Mvan_3489 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_2373|M.ulcerans_Agy99 ----------------MTQKNGETGTSTDSRDDVAEVVSRLIRFDTTNTG
MMAR_3123|M.marinum_M ----------------MTQKNGETGTSTDSRDDVAEVVSRLIRFDTTNTG
Mb2165c|M.bovis_AF2122/97 ----------------MTD---ETGASSDHSDDVAQVVSRLIRFDTTNSG
Rv2141c|M.tuberculosis_H37Rv ----------------MTD---ETGASSDHSDDVAQVVSRLIRFDTTNSG
MLBr_01288|M.leprae_Br4923 MQKTKVLRSLVVAVVGKVTVTVETDVLVNPSDDVVEVVSRLIRFDTTNTG
MAV_2350|M.avium_104 ----------------MTIQSGASEVTANPSDDVVEVVSTLIRFDTTNTG
Mflv_3037|M.gilvum_PYR-GCK ----------------MTGDAGGRSGGLTPSDEVVDLVSTLIRFDTSNTG
Mvan_3489|M.vanbaalenii_PYR-1 -------------------------------------MSTLIRFDTSNTG
MAB_2102|M.abscessus_ATCC_1997 ----------------MT------SVNPAPEDEVVDLVSTLIRFDTSNTG
MSMEG_4200|M.smegmatis_MC2_155 -----------------------MTVSAASADEVVDLVSALIRFDTSNTG
TH_2370|M.thermoresistible__bu -----------------------VTNPPQPADEVVDLVSTLIRFDTSNTG
:* ******:*:*
MUL_2373|M.ulcerans_Agy99 EPETTKGEAECARWVAEQLAEVGYQTEYVESGAPGRGNVFARLRGADSSR
MMAR_3123|M.marinum_M EPETTKGEAECARWVAEQLAEVGYQTEYVESGAPGRGNVFARLRGADSSR
Mb2165c|M.bovis_AF2122/97 EPGTTKGEAECARWVAEQLAEVGYQPEYVESGAPGRGNVFARLAGADSSR
Rv2141c|M.tuberculosis_H37Rv EPGTTKGEAECARWVAEQLAEVGYQPEYVESGAPGRGNVFARLAGADSSR
MLBr_01288|M.leprae_Br4923 DPETTRGEAECAQWVASQLAEVGYQPEYLESGAPGRGNVFARLAGEDSSR
MAV_2350|M.avium_104 EPETTKGEAECAQWVAEQLAAVGYAPHYVESGAPGRGNVFVRLPGADSSR
Mflv_3037|M.gilvum_PYR-GCK DPATTMGEAACARWVADQLAEVGYVCEYIEAGAPGRANVFARLEGADRSR
Mvan_3489|M.vanbaalenii_PYR-1 DPATTKGEADCARWVADQLAEVGYVTEYVEAGAPGRANVFARLEGADRSR
MAB_2102|M.abscessus_ATCC_1997 ELETTKGEAECARWIQQQLEEVGYTCEYVEAGAPGRGNLFARLPGASSDR
MSMEG_4200|M.smegmatis_MC2_155 DPATTKGEAECAHWVAQQLEEVGYETEYVESGAPGRGNVFARLRGADPSR
TH_2370|M.thermoresistible__bu DPATTRVEAEAAHWVADRLREVGYQTEYVEAGAPGRGNVFARLPGADPSR
: ** ** .*:*: .:* *** .*:*:*****.*:*.** * . .*
MUL_2373|M.ulcerans_Agy99 GALLIHGHLDVVPAEAADWSVHPFSGAIEDGYVWGRGAVDMKDMVGMMIV
MMAR_3123|M.marinum_M GALLIHGHLDVVPAEAADWSVHPFSGAIEDGYVWGRGAVDMKDMVGMMIV
Mb2165c|M.bovis_AF2122/97 GALLIHGHLDVVPAEPAEWSVHPFSGAIEDGYVWGRGAVDMKDMVGMMIV
Rv2141c|M.tuberculosis_H37Rv GALLIHGHLDVVPAEPAEWSVHPFSGAIEDGYVWGRGAVDMKDMVGMMIV
MLBr_01288|M.leprae_Br4923 GALLIHGHLDVVPAETAEWSVHPFSGAVEGGQVWGRGAIDMKDMVGMMIV
MAV_2350|M.avium_104 GALLIHGHLDVVPAEPTEWSVHPFSGAVKDGFVWGRGAVDMKDMVGMMIV
Mflv_3037|M.gilvum_PYR-GCK GALMLHGHLDVVPAEASDWSVHPFSGAVEDGYVWGRGAVDMKDMVGMILA
Mvan_3489|M.vanbaalenii_PYR-1 GALMLHGHLDVVPAEASDWSVHPFSGAVEDGYVWGRGAVDMKDMVGMIIA
MAB_2102|M.abscessus_ATCC_1997 GALLIHGHLDVVPAEAPDWSVHPFSGAVNDGYVWGRGAVDMKDMVGMMIA
MSMEG_4200|M.smegmatis_MC2_155 GALMVHGHLDVVPAEPADWSVHPFSGAVKDGYVWGRGAVDMKDMVGMTLA
TH_2370|M.thermoresistible__bu GALLVHGHLDVVPAEPADWSVHPFSGAVEDGYVWGRGAVDMKNMLGMMLA
***::**********..:*********::.* ******:***:*:** :.
MUL_2373|M.ulcerans_Agy99 VARHFRRAGIVPPRDLVFAFLADEEHGGHFGAHWLVDNRPDLFDGVTEAV
MMAR_3123|M.marinum_M VARHFRRAGIVPPRDLVFAFLADEEHGGHFGAHWLVDNRPDLFDGVTEAV
Mb2165c|M.bovis_AF2122/97 VARHLRQAAIVPPRDLVFAFVADEEHGGKYGSHWLVDNRPDLFDGITEAI
Rv2141c|M.tuberculosis_H37Rv VARHLRQAAIVPPRDLVFAFVADEEHGGKYGSHWLVDNRPDLFDGITEAI
MLBr_01288|M.leprae_Br4923 VARQLKQAGIAPPRDLVFAFVADEEHGGSYGSQWLVDNRPDLFDGVTEAI
MAV_2350|M.avium_104 VARHLKRAGIVPPRDLVFAFIADEEHGGTFGAQWLVDNRPELFAGVTEAI
Mflv_3037|M.gilvum_PYR-GCK VARHFKRSGIVPPRDLVFAFVSDEEAGGNYGCKWLVEHRPDLFEGVTEAV
Mvan_3489|M.vanbaalenii_PYR-1 VARHFKRSGIVPPRDLVFAFVSDEEAGGNFGCKWLVENRPDLFDGVTEAV
MAB_2102|M.abscessus_ATCC_1997 IARYFKRSGIVPPRDLVWAFVSDEENGGKWGSQWLVDNRPDLFEGITEAI
MSMEG_4200|M.smegmatis_MC2_155 VARHFKRAGIVPPRDLVFAFVADEEHGGTYGADWLVNNRPDLFEGVTEAI
TH_2370|M.thermoresistible__bu VARHFKRAGIVPPRDIVFAFLSDEEHGGTYGAQWLVDHRPDLFTGVTEAI
:** ::::.*.****:*:**::*** ** :*..***::**:** *:***:
MUL_2373|M.ulcerans_Agy99 GEVGGFSLTVPRRDGGERRLYLVETAEKGILWMRLTAQGRAGHGSMVHDH
MMAR_3123|M.marinum_M GEVGGFSLTVPRRDGGERRLYLVETAEKGILWMRLTAQGRAGHGSMVHDH
Mb2165c|M.bovis_AF2122/97 GEVGGFSLTVPRHDGGERRLYLIETAEKGIQWMRLTARGRAGHGSMVHDQ
Rv2141c|M.tuberculosis_H37Rv GEVGGFSLTVPRHDGGERRLYLIETAEKGIQWMRLTARGRAGHGSMVHDQ
MLBr_01288|M.leprae_Br4923 GEVGGFSLTVPCRNGGERRLYLIETAEKGMQWMRLTARGRAGHGSMVHNQ
MAV_2350|M.avium_104 GEVGGFSLTVPRRDGGERRLYLIETAEKGLSWMKLTARGPAGHGSMVHDQ
Mflv_3037|M.gilvum_PYR-GCK GEVGGFSLTVPRPDGGEKRLYLVETAEKAMLWMRLTARARAGHGSMVHDD
Mvan_3489|M.vanbaalenii_PYR-1 GEVGGFSLTVPRRDGGERRLYLVETAEKAMLWMRLTARGRAGHGSMVHDD
MAB_2102|M.abscessus_ATCC_1997 GEVGGFSLTVPRRQGGERRLYLLETAEKGIAWMRLTAKARAGHGSMVHED
MSMEG_4200|M.smegmatis_MC2_155 GEVGGFSLTVPRKDGGERRLYLIETAEKGLSWMRLTARGRAGHGSMVHDD
TH_2370|M.thermoresistible__bu GEVGGFSLTVPRKDGGERRLYLVETAEKGLAWMRLTARGRAGHGSMVNDD
*********** :***:****:*****.: **:***:. *******::.
MUL_2373|M.ulcerans_Agy99 NAVTTVAEAVARLGRHQFPLVITDTVAQFLHAVSEETGLDLDPDSPDLEG
MMAR_3123|M.marinum_M NAVTTVAEAVARLGRHQFPLVITDTVAQFLHAVSEETGLELDPGSPDLEG
Mb2165c|M.bovis_AF2122/97 NAVTAVCEAVARLGRHQFPLVCTDTVAQFLAVVGEETGLAFDLDSPDLAG
Rv2141c|M.tuberculosis_H37Rv NAVTAVCEAVARLGRHQFPLVCTDTVAQFLAVVGEETGLAFDLDSPDLAG
MLBr_01288|M.leprae_Br4923 NAVTAVAEAVARLGRHQFPLVTTDTVVQFLAAISEETGLEFDIGSPDLEG
MAV_2350|M.avium_104 NAVTAVAEAVARLGRHQFPLVLTDTVNQFLAAVSEETGLTFDTQSGDLRG
Mflv_3037|M.gilvum_PYR-GCK NAVTAVAEAVAKLGRHRFPIVLTESVEQFLTAVGEETGYAFDPASPDIEG
Mvan_3489|M.vanbaalenii_PYR-1 NAVTAVAEAVAKLGRHRFPIVLTESVEQFLTAVGEETGYAFDPASPDIEG
MAB_2102|M.abscessus_ATCC_1997 NAVTAVAEAVAKLGRHRFPLVLTEAVTQFLGAVAEETGYDFDPDSPDLEG
MSMEG_4200|M.smegmatis_MC2_155 NAVTAIAGAVDRLGRHEFPLVLSPAVEEFLTAVAEETGYTFDPNSPDLEG
TH_2370|M.thermoresistible__bu NAVTAVAEAVARLGRHRFPLVLTESVQQFLEAVAEETGYSFDVNSPDLEG
****::. ** :****.**:* : :* :** .:.**** :* * *: *
MUL_2373|M.ulcerans_Agy99 TLDKLGPIARMLKAVLRDTANPTMLKAGYKANVIPATAEAVVDCRVLPGR
MMAR_3123|M.marinum_M TLDKLGPIARMLKAVLRDTANPTMLKAGYKANVIPATAEAVVDCRVLPGR
Mb2165c|M.bovis_AF2122/97 TIDKLGPMARMLKAVLHDTANPTMLKAGYKANVVPATAEAVVDCRVLPGR
Rv2141c|M.tuberculosis_H37Rv TIDKLGPMARMLKAVLHDTANPTMLKAGYKANVVPATAEAVVDCRVLPGR
MLBr_01288|M.leprae_Br4923 AIEKLGPMARMLKAVLYDTANPTVLKAGYKVNVVPATAEAMVDCRILPGR
MAV_2350|M.avium_104 VVEKLGPMARMLKAVLHDTANPTMLKAGYKANVVPAIAEAVVDCRILPGR
Mflv_3037|M.gilvum_PYR-GCK TVAKLGGIARIVGATLRDTANPTMLKAGYKANVIPATAEAVIDCRVLPGR
Mvan_3489|M.vanbaalenii_PYR-1 TVAKLGGIARIVGATLRDTANPTMLNAGYKANVIPATAEAVIDCRVLPGR
MAB_2102|M.abscessus_ATCC_1997 TVAKLGSIGKIVGATLRDTANPTMLKAGYKANVIPATAEAVIDCRVLPGR
MSMEG_4200|M.smegmatis_MC2_155 TIAKLGGVARIVSATLRDTANPTMLKAGYKANVIPAVAEAMIDCRVLPGR
TH_2370|M.thermoresistible__bu TIAKLGGVARIVGATLRDTANPTMLTAGYKANVIPQTAEAIVDCRVLPGR
.: *** :.::: *.* ******:*.****.**:* ***::***:****
MUL_2373|M.ulcerans_Agy99 QAAFEAEVDELIGPDVSREWISELASYETSFDGDLVDAMNAAVLSVDPDG
MMAR_3123|M.marinum_M QAAFEAEVDELIGPDVSREWISELASYETSFDGDLVDAMNAAVLSVDPDG
Mb2165c|M.bovis_AF2122/97 RAAFEAEVDALIGPDVTREWVSDLPSYETTFDGDLVAAMNAAVLAVDPDG
Rv2141c|M.tuberculosis_H37Rv RAAFEAEVDALIGPDVTREWVSDLPSYETTFDGDLVAAMNAAVLAVDPDG
MLBr_01288|M.leprae_Br4923 QAAFEAAIDELIGPDVTREWIKDLPPYETAFDGDLVDAMNAAVLAVDPDG
MAV_2350|M.avium_104 KAAFEAEIDELIGPDVTREWIKDFSSYETGFDGDLVDAMNDAVLALDPDA
Mflv_3037|M.gilvum_PYR-GCK LADFEREVDELIGPDVKREWITNLPSYETPFDGELLDAMNTAILANDPDA
Mvan_3489|M.vanbaalenii_PYR-1 LADFEREVDELIGPDVKREWITNLPSYETPFDGELLEAMNNAILAADPDA
MAB_2102|M.abscessus_ATCC_1997 LEAFEREVDEIIGPDVEREWVQLLPAYETTFDGDLVDAMNAAVLEFDPEA
MSMEG_4200|M.smegmatis_MC2_155 KEAFEREVDELIGPDVTRSWERDLPSYETSFDGDLVDAMNASVLTLDPEA
TH_2370|M.thermoresistible__bu RAAFEREVDELIGPDVERSWERDLPAVETTFDGELVDAMNASLLAVDPEA
** :* :***** *.* :.. ** ***:*: *** ::* **:.
MUL_2373|M.ulcerans_Agy99 RTVPYMLSGGTDAKAFARLGIRCFGFSPLRLPPELDFATLFHGVDERVPI
MMAR_3123|M.marinum_M RTVPYMLSGGTDAKAFARLGIRCFGFSPLRLPPELDFAALFHGVDERVPI
Mb2165c|M.bovis_AF2122/97 RTVPYMLSGGTDAKAFARLGIRCFGFSPLRLPPDLDFTSLFHGVDERVPI
Rv2141c|M.tuberculosis_H37Rv RTVPYMLSGGTDAKAFARLGIRCFGFSPLRLPPDLDFTSLFHGVDERVPI
MLBr_01288|M.leprae_Br4923 RTVPYMASGGTDAKAFARLGIRCFGFTPLRLPPELDFTALFHGVDERVSI
MAV_2350|M.avium_104 RTVPYMLSGGTDAKSFARLGIRCFGFSPLRLPPDLDFTALFHGVDERVPI
Mflv_3037|M.gilvum_PYR-GCK RTVPYMLSGGTDAKHFARLGIRCFGFAPLRLPPDLDFAALFHGVDERVPV
Mvan_3489|M.vanbaalenii_PYR-1 RTVPYMMSGGTDAKHFARLGIRCFGFAPLRLPPDLDFAALFHGVDERVPV
MAB_2102|M.abscessus_ATCC_1997 RTVPYMLSGGTDAKAFARLGIRCFGFAPLKLPPELDFASLFHGVDERVPV
MSMEG_4200|M.smegmatis_MC2_155 RIVPYMLSAGTDAKSFQRLGIRCFGFAPLRLPPDLDFAALFHGVDERVPV
TH_2370|M.thermoresistible__bu RTVPYMMSGGTDAKAFQRLGIRCFGFVPLRLPPELDFSALFHGVDERVPV
* **** *.***** * ********* **:***:***::*********.:
MUL_2373|M.ulcerans_Agy99 DALRFGTEVLTHFLTHC
MMAR_3123|M.marinum_M DALRFGTEVLTHFLTHC
Mb2165c|M.bovis_AF2122/97 DGLRFGTEVLTHLLTHC
Rv2141c|M.tuberculosis_H37Rv DGLRFGTEVLTHLLTHC
MLBr_01288|M.leprae_Br4923 DALKFGIDVLAHFLTHC
MAV_2350|M.avium_104 DALRFGTDVLTHFLTHC
Mflv_3037|M.gilvum_PYR-GCK DALTFGAQVLEHFLLHS
Mvan_3489|M.vanbaalenii_PYR-1 DALTFGAQVLENFLLHS
MAB_2102|M.abscessus_ATCC_1997 DALTFGTKVLQHFLLNH
MSMEG_4200|M.smegmatis_MC2_155 DALQFGAGVLEHFLQNC
TH_2370|M.thermoresistible__bu DALEFGARVLDHFLQHC
*.* ** ** ::* :