For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLWGYHEVPEDQLATDFPAPDDPGMIVMHGISFSSTCPHHLLPFTGTATIAYRPHPGQRIVGLSKLARLV DGYAARLQVQERIGQQAVDGIMRRLNPSGAMCIITARHDCMRLRGVRDPAAEATTEARKGIWSDHEMALI HRLHGGAR
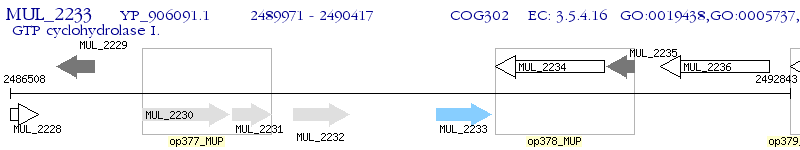
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2233 | folE_1 | - | 100% (148) | GTP cyclohydrolase I FolE_1 |
| M. ulcerans Agy99 | MUL_4191 | folE | 7e-25 | 44.09% (127) | GTP cyclohydrolase I |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3639c | folE | 1e-25 | 43.18% (132) | GTP cyclohydrolase I |
| M. gilvum PYR-GCK | Mflv_1417 | folE | 1e-26 | 46.46% (127) | GTP cyclohydrolase I |
| M. tuberculosis H37Rv | Rv3609c | folE | 1e-25 | 43.18% (132) | GTP cyclohydrolase I |
| M. leprae Br4923 | MLBr_00223 | folE | 2e-24 | 43.31% (127) | GTP cyclohydrolase I |
| M. abscessus ATCC 19977 | MAB_0534 | - | 3e-25 | 44.09% (127) | GTP cyclohydrolase I (FolE) |
| M. marinum M | MMAR_2992 | folE_1 | 6e-84 | 97.97% (148) | GTP cyclohydrolase I FolE_1 |
| M. avium 104 | MAV_0543 | folE | 8e-26 | 43.18% (132) | GTP cyclohydrolase I |
| M. smegmatis MC2 155 | MSMEG_6104 | folE | 8e-26 | 43.85% (130) | GTP cyclohydrolase I |
| M. thermoresistible (build 8) | TH_0990 | folE | 2e-26 | 46.46% (127) | GTP CYCLOHYDROLASE I FOLE (GTP-CH-I) |
| M. vanbaalenii PYR-1 | Mvan_5371 | folE | 3e-26 | 46.46% (127) | GTP cyclohydrolase I |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3639c|M.bovis_AF2122/97 ---------------------MSQLDSR---SASARIRVFDQQ-------
Rv3609c|M.tuberculosis_H37Rv ---------------------MSQLDSR---SASARIRVFDQQ-------
MAV_0543|M.avium_104 ---------------------MAGNGSAPD-TATHQVRQFDQA-------
MLBr_00223|M.leprae_Br4923 ---------------------MALLDLGLESTAVPRIRVFDQQ-------
MAB_0534|M.abscessus_ATCC_1997 ---------------------MNSPETAEY-NGHPTGHVFDQA-------
Mflv_1417|M.gilvum_PYR-GCK ---------------------MTRSHNHSA-TLT--TPDFDQA-------
Mvan_5371|M.vanbaalenii_PYR-1 ---------------------MTRSHNHSA-TIT--TARFDQA-------
TH_0990|M.thermoresistible__bu ---------------------MTRAHHNSA-TLTTKTPVFDQP-------
MSMEG_6104|M.smegmatis_MC2_155 ---------------------MTQSLRGHQ-NNN-VRRVFDQP-------
MUL_2233|M.ulcerans_Agy99 --------------------------------------------------
MMAR_2992|M.marinum_M MPSAPHQGRYQMPPLRPSAWQMTNDGVAYGAASYHPNSGYPHSGRRPMNS
Mb3639c|M.bovis_AF2122/97 ----------RAEAAVRELLYAIGEDPDRDGLVATPSRVARSYREMFAGL
Rv3609c|M.tuberculosis_H37Rv ----------RAEAAVRELLYAIGEDPDRDGLVATPSRVARSYREMFAGL
MAV_0543|M.avium_104 ----------RAEAAVRELLFAIGENPDRHGLAETPARVARAYREMFAGL
MLBr_00223|M.leprae_Br4923 ----------RAEAAIRELLYAIGEDPDREGLADTPARVARACRELFSGL
MAB_0534|M.abscessus_ATCC_1997 ----------RAEAAVRELLYAVGEDPDRHGLADTPARVARAYREIFAGL
Mflv_1417|M.gilvum_PYR-GCK ----------RAEAAVRELLIAVGEDPDREGLLDTPARVARSYREIFAGL
Mvan_5371|M.vanbaalenii_PYR-1 ----------RAEAAVRELLIAVGEDPDREGLRDTPARVARAYQEIFAGL
TH_0990|M.thermoresistible__bu ----------RAEAAIRELLLAVGEDPDRDGLRDTPARVARAYKEVFAGL
MSMEG_6104|M.smegmatis_MC2_155 ----------RAEAAVRELLIAIGEDPEREGLVDTPARVARAYKELMAGL
MUL_2233|M.ulcerans_Agy99 ---------------------------------------------MLWGY
MMAR_2992|M.marinum_M PAWYDNNPDLAVECAVQQLLDALGVD-DGEHTAGTPARVARAWREMLWGY
:: *
Mb3639c|M.bovis_AF2122/97 YTDPDSVLNTMFDEDHD-ELVLVKEIPMYSTCEHHLVAFHGVAHVGYIPG
Rv3609c|M.tuberculosis_H37Rv YTDPDSVLNTMFDEDHD-ELVLVKEIPMYSTCEHHLVAFHGVAHVGYIPG
MAV_0543|M.avium_104 YTDPDSVLNTMFDEEHD-ELVLVKEIPLYSTCEHHLVSFHGVAHVGYIPG
MLBr_00223|M.leprae_Br4923 YTDPQTVLNTMFDEEHN-ELVIVKEIPMYSTCEHHLVSFHGVAHIGYLPG
MAB_0534|M.abscessus_ATCC_1997 YTDPDTVLNTTFDEQHD-ELVLVKSIPMYSTCEHHLVSFHGVAHVGYIPG
Mflv_1417|M.gilvum_PYR-GCK YTDPDEVLTTMFDEQHD-EMVLVKDIPMYSTCEHHLVSFHGVAHVGYIPG
Mvan_5371|M.vanbaalenii_PYR-1 YTDPDEVLKTMFDEQHD-EMVLVKDIPMYSTCEHHLVSFHGVAHVGYIPG
TH_0990|M.thermoresistible__bu YTDPDAVLDTTFDEQHD-ELVLVRQIPLYSTCEHHLVSFHGFAHVGYIPG
MSMEG_6104|M.smegmatis_MC2_155 HTDPDSVLNTTFDEGHD-ELVLVKQIPMYSTCEHHLVSFHGVAHVGYIPG
MUL_2233|M.ulcerans_Agy99 HEVPEDQLATDFPAPDDPGMIVMHGISFSSTCPHHLLPFTGTATIAYRPH
MMAR_2992|M.marinum_M HEVPEDQLATDFPAPDDPGLIVMHGISFSSTCAHHLLPFTGTATIAYRPH
: *: * * * .: ::::: *.: *** ***:.* * * :.* *
Mb3639c|M.bovis_AF2122/97 DDGRVTGLSKIARLVDLYAKRPQVQERLTSQIADALMKKLDPRGVIVVIE
Rv3609c|M.tuberculosis_H37Rv DDGRVTGLSKIARLVDLYAKRPQVQERLTSQIADALMKKLDPRGVIVVIE
MAV_0543|M.avium_104 NDGRVTGLSKIARLVDLYAKRPQVQERLTSQIADALVKKLNPRGVIVVVE
MLBr_00223|M.leprae_Br4923 ADGRVTGLSKIARLVDLYAKRPQVQERLTSQIADALVSKLDPRGVIIVVE
MAB_0534|M.abscessus_ATCC_1997 QHGRVTGLSKIARLVDLYAKRPQVQERLTAQIADALVRKLEPRGVIVVVE
Mflv_1417|M.gilvum_PYR-GCK VDGRVTGLSKLARVVDLYAKRPQVQERLTSQIADALMRKLDPRGAIVVIE
Mvan_5371|M.vanbaalenii_PYR-1 VDGRVTGLSKLARVVDLYAKRPQVQERLTGQIADALMRRLDPRGVIVVIE
TH_0990|M.thermoresistible__bu DDGRVTGLSKIARLVDLYAKRPQVQERLTAQIADALMRKLNPRGAIVVVE
MSMEG_6104|M.smegmatis_MC2_155 VDGRVTGLSKIARLVDLYSKRPQVQERLTAQIADALMRKLDPRGVIVVVE
MUL_2233|M.ulcerans_Agy99 PGQRIVGLSKLARLVDGYAARLQVQERIGQQAVDGIMRRLNPSGAMCIIT
MMAR_2992|M.marinum_M PGQRIVGLSKLARLVHGYAARLQVQERIGQQAVDGIMRRLNPSGAMCIIT
*:.****:**:*. *: * *****: * .*.:: :*:* *.: ::
Mb3639c|M.bovis_AF2122/97 AEHLCMAMRGVRKPGSVTTTSAVRGLFKTNAASRAEALDLILRK
Rv3609c|M.tuberculosis_H37Rv AEHLCMAMRGVRKPGSVTTTSAVRGLFKTNAASRAEALDLILRK
MAV_0543|M.avium_104 AEHLCMAMRGVRKPGAVTTTSAVRGLFKTNAASRAEALDLILRK
MLBr_00223|M.leprae_Br4923 AEHLCMAMRGVRKPGAITTTSAVRGQFKTDAASRAEALGLILRK
MAB_0534|M.abscessus_ATCC_1997 AEHLCMAMRGVRKPGATTTTSAVRGQFKRDAASRAEVLDLMMRT
Mflv_1417|M.gilvum_PYR-GCK AEHLCMAMRGIRKPGAVTTTSAVRGQFKTDKASRAEALDLILRK
Mvan_5371|M.vanbaalenii_PYR-1 AEHLCMAMRGIRKPGAVTTTSAVRGQFKTDKASRAEALDLILRK
TH_0990|M.thermoresistible__bu AEHLCMAMRGVRKPGALTTTSAVRGQFKTDNASRAEALELILRK
MSMEG_6104|M.smegmatis_MC2_155 AEHLCMAMRGVRKPGAVTTTSAVRGQFKTDKASRAEALELILRK
MUL_2233|M.ulcerans_Agy99 ARHDCMRLRGVRDPAAEATTEARKGIWSDHEMALIHRLHGGAR-
MMAR_2992|M.marinum_M ARHDCMRLRGVRDPAAEATTEARKGIWSDHEMALIHRLHGGAR-
*.* ** :**:*.*.: :**.* :* :. . : . * *