For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRSHNHSATITTARFDQARAEAAVRELLIAVGEDPDREGLRDTPARVARAYQEIFAGLYTDPDEVLKTMF DEQHDEMVLVKDIPMYSTCEHHLVSFHGVAHVGYIPGVDGRVTGLSKLARVVDLYAKRPQVQERLTGQIA DALMRRLDPRGVIVVIEAEHLCMAMRGIRKPGAVTTTSAVRGQFKTDKASRAEALDLILRK*
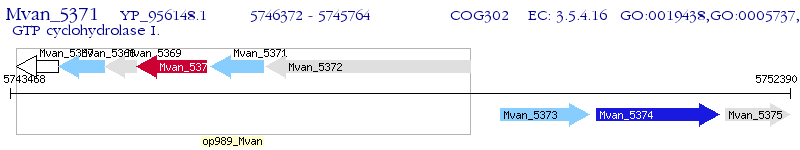
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5371 | folE | - | 100% (202) | GTP cyclohydrolase I |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3639c | folE | 1e-91 | 80.69% (202) | GTP cyclohydrolase I |
| M. gilvum PYR-GCK | Mflv_1417 | folE | 1e-107 | 95.05% (202) | GTP cyclohydrolase I |
| M. tuberculosis H37Rv | Rv3609c | folE | 1e-91 | 80.69% (202) | GTP cyclohydrolase I |
| M. leprae Br4923 | MLBr_00223 | folE | 1e-90 | 82.89% (187) | GTP cyclohydrolase I |
| M. abscessus ATCC 19977 | MAB_0534 | - | 7e-91 | 86.56% (186) | GTP cyclohydrolase I (FolE) |
| M. marinum M | MMAR_5112 | folE | 9e-96 | 83.66% (202) | GTP cyclohydrolase I FolE |
| M. avium 104 | MAV_0543 | folE | 4e-92 | 83.08% (195) | GTP cyclohydrolase I |
| M. smegmatis MC2 155 | MSMEG_6104 | folE | 1e-93 | 83.25% (203) | GTP cyclohydrolase I |
| M. thermoresistible (build 8) | TH_0990 | folE | 7e-97 | 83.33% (204) | GTP CYCLOHYDROLASE I FOLE (GTP-CH-I) |
| M. ulcerans Agy99 | MUL_4191 | folE | 1e-95 | 83.17% (202) | GTP cyclohydrolase I |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3639c|M.bovis_AF2122/97 MSQLDSR---SASARIRVFDQQRAEAAVRELLYAIGEDPDRDGLVATPSR
Rv3609c|M.tuberculosis_H37Rv MSQLDSR---SASARIRVFDQQRAEAAVRELLYAIGEDPDRDGLVATPSR
MAV_0543|M.avium_104 MAGNGSAPD-TATHQVRQFDQARAEAAVRELLFAIGENPDRHGLAETPAR
MMAR_5112|M.marinum_M MTQLDSR---TEATTTRAFDQPRAEAAVRELLLAIGEDPDRGGLRDTPAR
MUL_4191|M.ulcerans_Agy99 MTQLDSR---TEATTTRAFDQPRAEAAVRELLLAIGEDPDRGGLRDTPAR
MLBr_00223|M.leprae_Br4923 MALLDLGLESTAVPRIRVFDQQRAEAAIRELLYAIGEDPDREGLADTPAR
MAB_0534|M.abscessus_ATCC_1997 MNSPETAEY-NGHPTGHVFDQARAEAAVRELLYAVGEDPDRHGLADTPAR
Mvan_5371|M.vanbaalenii_PYR-1 MTRSHNHSA-TIT--TARFDQARAEAAVRELLIAVGEDPDREGLRDTPAR
Mflv_1417|M.gilvum_PYR-GCK MTRSHNHSA-TLT--TPDFDQARAEAAVRELLIAVGEDPDREGLLDTPAR
TH_0990|M.thermoresistible__bu MTRAHHNSA-TLTTKTPVFDQPRAEAAIRELLLAVGEDPDRDGLRDTPAR
MSMEG_6104|M.smegmatis_MC2_155 MTQSLRGHQ-NNN-VRRVFDQPRAEAAVRELLIAIGEDPEREGLVDTPAR
* . *** *****:**** *:**:*:* ** **:*
Mb3639c|M.bovis_AF2122/97 VARSYREMFAGLYTDPDSVLNTMFDEDHDELVLVKEIPMYSTCEHHLVAF
Rv3609c|M.tuberculosis_H37Rv VARSYREMFAGLYTDPDSVLNTMFDEDHDELVLVKEIPMYSTCEHHLVAF
MAV_0543|M.avium_104 VARAYREMFAGLYTDPDSVLNTMFDEEHDELVLVKEIPLYSTCEHHLVSF
MMAR_5112|M.marinum_M VARAYREIFAGLYTDPDAVLNTMFDEDHDELVLIKEIPLYSTCEHHLVSF
MUL_4191|M.ulcerans_Agy99 VARAYREIFAGLYTDPDAVLNTMFDEDHDELVLIKEIPLYSTCEHHLVSF
MLBr_00223|M.leprae_Br4923 VARACRELFSGLYTDPQTVLNTMFDEEHNELVIVKEIPMYSTCEHHLVSF
MAB_0534|M.abscessus_ATCC_1997 VARAYREIFAGLYTDPDTVLNTTFDEQHDELVLVKSIPMYSTCEHHLVSF
Mvan_5371|M.vanbaalenii_PYR-1 VARAYQEIFAGLYTDPDEVLKTMFDEQHDEMVLVKDIPMYSTCEHHLVSF
Mflv_1417|M.gilvum_PYR-GCK VARSYREIFAGLYTDPDEVLTTMFDEQHDEMVLVKDIPMYSTCEHHLVSF
TH_0990|M.thermoresistible__bu VARAYKEVFAGLYTDPDAVLDTTFDEQHDELVLVRQIPLYSTCEHHLVSF
MSMEG_6104|M.smegmatis_MC2_155 VARAYKELMAGLHTDPDSVLNTTFDEGHDELVLVKQIPMYSTCEHHLVSF
***: :*:::**:***: ** * *** *:*:*:::.**:*********:*
Mb3639c|M.bovis_AF2122/97 HGVAHVGYIPGDDGRVTGLSKIARLVDLYAKRPQVQERLTSQIADALMKK
Rv3609c|M.tuberculosis_H37Rv HGVAHVGYIPGDDGRVTGLSKIARLVDLYAKRPQVQERLTSQIADALMKK
MAV_0543|M.avium_104 HGVAHVGYIPGNDGRVTGLSKIARLVDLYAKRPQVQERLTSQIADALVKK
MMAR_5112|M.marinum_M HGVAHVGYIPGRDGRVTGLSKIARLVDLYAKRPQVQERLTSQIADALVKR
MUL_4191|M.ulcerans_Agy99 HGVAHVGYIPGRDGRVTGLSKIARLVDLYAKRPQVQERLTSQIADALVKR
MLBr_00223|M.leprae_Br4923 HGVAHIGYLPGADGRVTGLSKIARLVDLYAKRPQVQERLTSQIADALVSK
MAB_0534|M.abscessus_ATCC_1997 HGVAHVGYIPGQHGRVTGLSKIARLVDLYAKRPQVQERLTAQIADALVRK
Mvan_5371|M.vanbaalenii_PYR-1 HGVAHVGYIPGVDGRVTGLSKLARVVDLYAKRPQVQERLTGQIADALMRR
Mflv_1417|M.gilvum_PYR-GCK HGVAHVGYIPGVDGRVTGLSKLARVVDLYAKRPQVQERLTSQIADALMRK
TH_0990|M.thermoresistible__bu HGFAHVGYIPGDDGRVTGLSKIARLVDLYAKRPQVQERLTAQIADALMRK
MSMEG_6104|M.smegmatis_MC2_155 HGVAHVGYIPGVDGRVTGLSKIARLVDLYSKRPQVQERLTAQIADALMRK
**.**:**:** .********:**:****:**********.******: :
Mb3639c|M.bovis_AF2122/97 LDPRGVIVVIEAEHLCMAMRGVRKPGSVTTTSAVRGLFKTNAASRAEALD
Rv3609c|M.tuberculosis_H37Rv LDPRGVIVVIEAEHLCMAMRGVRKPGSVTTTSAVRGLFKTNAASRAEALD
MAV_0543|M.avium_104 LNPRGVIVVVEAEHLCMAMRGVRKPGAVTTTSAVRGLFKTNAASRAEALD
MMAR_5112|M.marinum_M LGPRGVIVVVEAEHLCMAMRGVRKPGAVTTTSAVRGQFKTDAASRAEALD
MUL_4191|M.ulcerans_Agy99 LGPRGVLVVVEAEHLCMAMRGVRKPGAVTTTSAVRGQFKTDAASRAEALD
MLBr_00223|M.leprae_Br4923 LDPRGVIIVVEAEHLCMAMRGVRKPGAITTTSAVRGQFKTDAASRAEALG
MAB_0534|M.abscessus_ATCC_1997 LEPRGVIVVVEAEHLCMAMRGVRKPGATTTTSAVRGQFKRDAASRAEVLD
Mvan_5371|M.vanbaalenii_PYR-1 LDPRGVIVVIEAEHLCMAMRGIRKPGAVTTTSAVRGQFKTDKASRAEALD
Mflv_1417|M.gilvum_PYR-GCK LDPRGAIVVIEAEHLCMAMRGIRKPGAVTTTSAVRGQFKTDKASRAEALD
TH_0990|M.thermoresistible__bu LNPRGAIVVVEAEHLCMAMRGVRKPGALTTTSAVRGQFKTDNASRAEALE
MSMEG_6104|M.smegmatis_MC2_155 LDPRGVIVVVEAEHLCMAMRGVRKPGAVTTTSAVRGQFKTDKASRAEALE
* ***.::*:***********:****: ******** ** : *****.*
Mb3639c|M.bovis_AF2122/97 LILRK
Rv3609c|M.tuberculosis_H37Rv LILRK
MAV_0543|M.avium_104 LILRK
MMAR_5112|M.marinum_M LILRK
MUL_4191|M.ulcerans_Agy99 LILRK
MLBr_00223|M.leprae_Br4923 LILRK
MAB_0534|M.abscessus_ATCC_1997 LMMRT
Mvan_5371|M.vanbaalenii_PYR-1 LILRK
Mflv_1417|M.gilvum_PYR-GCK LILRK
TH_0990|M.thermoresistible__bu LILRK
MSMEG_6104|M.smegmatis_MC2_155 LILRK
*::*.