For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MHVGYNTNSLADHPIAEALALISEEGYSAVALTVGYPHVRPMDSDLGAQLDALGCLLDTYGLTVAIETDA RYLLDPHNKHKPSPVDVAAQPRIEFLQRAIDIAAELGATCVSLWSGYSVSYSDAATTRDLLLSRLSRLVE YADRKSVMLGFEPEPGMFVETVQQALGLCSELGDPARLGITLDVGHCVMTEPAGAEAAIAELGDKLVNVH LDDMTPLKHEHLEFGYGALDLGAVMDALQVAGFAGVASVELPRHAHDAPKVARRSMNSIKLPQLPLQVRT WIAEAAAVLRRDPEKVLELFSGAQCQMPASSDEATVLARGRLVATLVAETPDVTAGPLIDKLYRWGDSDE RLGVFCGLETAASEETLGPDATRVGVGLAEDALRCNDPRLVAAALGGFGARFLAQHRWRDGVMKLVFMGV PLRGVSSLCSRADTELGRMATDYASERAAAGRMIPADAQLLQRLCATEAEALK
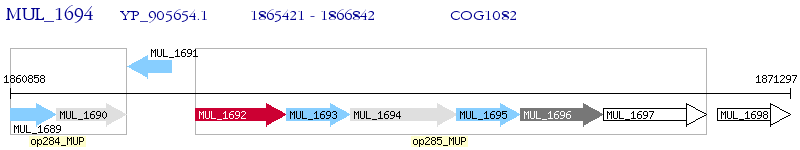
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1694 | - | - | 100% (473) | sugar phosphate isomerases/epimerases |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2510 | - | 0.0 | 98.31% (473) | sugar phosphate isomerase/epimerase |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6021 | xylA | 4e-08 | 30.41% (171) | xylose isomerase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5294 | - | 9e-09 | 22.71% (339) | xylose isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6021|M.smegmatis_MC2_155 -----MTVLESNVSTTEPLTPKREDRFSFGLWTIGWNGNDPFG-------
Mvan_5294|M.vanbaalenii_PYR-1 -----MTVLESSRPVPDALSPRREDKFSFGLWTVGWPGVDPFG-------
MUL_1694|M.ulcerans_Agy99 MHVGYNTNSLADHPIAEALALISEEGYSAVALTVGYPHVRPMDSDLGAQL
MMAR_2510|M.marinum_M MHVGYNTNSLADHPIAEALALIAEEGYSAVALTVGYPHVRPMDSDLGAQL
* :. . .:.*: *: :* *:*: *:.
MSMEG_6021|M.smegmatis_MC2_155 ------------------------------------VATRPALDVVE-AV
Mvan_5294|M.vanbaalenii_PYR-1 ------------------------------------VATRPALDAIE-AV
MUL_1694|M.ulcerans_Agy99 DALGCLLDTYGLTVAIETDARYLLDPHNKHKPSPVDVAAQPRIEFLQRAI
MMAR_2510|M.marinum_M DALGCLLDTYGLTVAIETGARYLLDPHNKHKPSLVDVAAQPRIEFLQRAI
**::* :: :: *:
MSMEG_6021|M.smegmatis_MC2_155 EKLAELGAYGLTFHDDDLFAFGSSDTERRRMVDRLTSALSANGMVVPMVT
Mvan_5294|M.vanbaalenii_PYR-1 DRLAALGAYGLTFHDDDLFAFGSSDGERRRAIDRLTSALSANGLVVPMVT
MUL_1694|M.ulcerans_Agy99 DIAAELGATCVSLWSGYSVSYSDAATTRDLLLSRLSRLVEYADRKSVMLG
MMAR_2510|M.marinum_M DIAAELGATCVSLWSGYSVSYSDAATTRDLLLSRLSRLVEYADRKSVILG
: * *** ::: .. .::..: * :.**: :. . ::
MSMEG_6021|M.smegmatis_MC2_155 TN----LFTQPVFKDGGFTSNDRTVRRFALRKVLRNIDLAIELGAETFVL
Mvan_5294|M.vanbaalenii_PYR-1 TN----LFTQPVFKDGGFTSNDRAVRRFALRKVLRNLDLAAELGAQTFVM
MUL_1694|M.ulcerans_Agy99 FEPEPGMFVETVQQALGLCSELGDPARLGITLDVGHCVMTEPAGAEAAIA
MMAR_2510|M.marinum_M FEPEPGMFVETVQQALGLCSELGDPARLGITLDVGHCVMTEPAGAEAAIA
: :*.:.* : *: *: *:.: : : :: **:: :
MSMEG_6021|M.smegmatis_MC2_155 WGGREGSEYDSAKDVQAALARYREAMDLLCQYVIDQGSGLR-FAIEPKPN
Mvan_5294|M.vanbaalenii_PYR-1 WGGREGGEYDSAKDVQAALERYREALDVLCQYVIDQGSGIR-FAIEPKPN
MUL_1694|M.ulcerans_Agy99 ELGDKLVNVHLDDMTPLKHEHLEFGYGALDLGAVMDALQVAGFAGVASVE
MMAR_2510|M.marinum_M ELGDKLVNVHLDDMTPLKHEHLEFGYGALDLGAVMDALQVAGFAGVASVE
* : : . . . : . . . * .: :. : ** .. :
MSMEG_6021|M.smegmatis_MC2_155 EPRGDILLPTVGHALAFIDTLARPEMVGVNPETGHEQMSGLNFMHGIAQA
Mvan_5294|M.vanbaalenii_PYR-1 EPRGDILLPTVGHALAFIETLAHPEMVGVNPETGHEQMSGLNFTHGIAQA
MUL_1694|M.ulcerans_Agy99 LPRHAHDAPKV--ARRSMNSIKLPQLPLQVRTWIAEAAAVLRRDPEKVLE
MMAR_2510|M.marinum_M LPRHAHDAPKV--ARRSMNSIKLAQLPLQVRTWIAEAAAVLRRDPEKVLE
** *.* * :::: .:: * : *. .
MSMEG_6021|M.smegmatis_MC2_155 LYSGKLFHIDLNGQRGIKFDQ-----DLVFGHGDLANAFALVDLLEHGGP
Mvan_5294|M.vanbaalenii_PYR-1 LYSGKLFHIDLNGQRGIKFDQ-----DLVFGHGDLVNAFSLVDLLEHGGP
MUL_1694|M.ulcerans_Agy99 LFSGAQCQMPASSDEATVLARGRLVATLVAETPDVTAGPLIDKLYRWGDS
MMAR_2510|M.marinum_M LFSGAQRQMPASSDEATVLARGRLVATLVAETPDVTAGPLIDKLYRWGDS
*:** :: ..:.. : : ** *:. . : .* . *..
MSMEG_6021|M.smegmatis_MC2_155 DGTP-AYEGPRHFDYKPSRTEDIDGVWASAAANMR-------MYLLLKQR
Mvan_5294|M.vanbaalenii_PYR-1 DGGP-AYDGPRHFDYKPSRTEDAEGVWASAAANMR-------MYLLLRQR
MUL_1694|M.ulcerans_Agy99 DERLGVFCGLETAASEETLGPDATRVGVGLAEDALRCNDPRLVAAALGGF
MMAR_2510|M.marinum_M DERLGVFCGLETAASEETLGPDATRVGVGLAEDALRCNDPRLVAAALGGF
* .: * . : : * * .. * : : *
MSMEG_6021|M.smegmatis_MC2_155 AEAFRADPAVREAMAAAKVAELRQPTLAPGETYHDLLADRSAFEEFDSEA
Mvan_5294|M.vanbaalenii_PYR-1 AAAFRADPDVREAMVAAKVAELRKPTLAAGETYADLLNDRSAYEEFDTGA
MUL_1694|M.ulcerans_Agy99 GARFLAQHRWRDGVMKLVFMGVPLRGVSSLCSRADTELGRMATDYASERA
MMAR_2510|M.marinum_M GARFLAQHRWRDGVMKLVFMGVPLRGVSGLRSRADTELGRMATDYASERA
. * *: *:.: . : :: : * .* * : . *
MSMEG_6021|M.smegmatis_MC2_155 YFGGKG-CGFVALQQLAIEHLMGAR
Mvan_5294|M.vanbaalenii_PYR-1 YFGGKG-CGFVALHQLAVEHLMGAR
MUL_1694|M.ulcerans_Agy99 AAGRMIPADAQLLQRLCATEAEALK
MMAR_2510|M.marinum_M AAGRMIPADAQLLQRLCATEAEALK
* .. *::*. . . :