For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IVTASVLCKTTDTVRGIALALRTAARQMSRLRKVSTVAGITLLVAVALLVPLPTAVQMRDWASSVGPWLP LVFLLVHIVVTVPPFPRTAFTVAGGLLFGPVLGIVIAVTASTASAVIALLLVRAAGWRLNRLVPHGAVDR LDERLRERGWLAILSLRLIPAVPFAALNYGAGASAVRVVPYTLSTLAGLLPGTVAVVILGSALAGDGSPL LVLVSVCTAALGMSGLVFEVRQYRRHHRHQAPSDHGPVLEPACEPLGV*
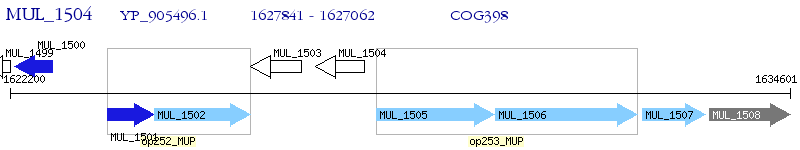
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1504 | - | - | 100% (259) | hypothetical protein MUL_1504 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1528c | - | 1e-95 | 69.72% (251) | hypothetical protein Mb1528c |
| M. gilvum PYR-GCK | Mflv_3651 | - | 7e-76 | 62.33% (223) | hypothetical protein Mflv_3651 |
| M. tuberculosis H37Rv | Rv1491c | - | 1e-95 | 69.72% (251) | hypothetical protein Rv1491c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2713 | - | 3e-69 | 58.02% (212) | hypothetical protein MAB_2713 |
| M. marinum M | MMAR_2301 | - | 1e-142 | 100.00% (257) | hypothetical protein MMAR_2301 |
| M. avium 104 | MAV_3279 | - | 2e-89 | 66.67% (252) | hypothetical protein MAV_3279 |
| M. smegmatis MC2 155 | MSMEG_3157 | - | 4e-85 | 68.56% (229) | hypothetical protein MSMEG_3157 |
| M. thermoresistible (build 8) | TH_0612 | - | 3e-60 | 61.11% (180) | CONSERVED MEMBRANE PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_2761 | - | 6e-79 | 61.02% (236) | hypothetical protein Mvan_2761 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_1504|M.ulcerans_Agy99 -----------------------MIVTASVLCKTTDTVRGIALALRTAAR
MMAR_2301|M.marinum_M -------------------------MTASVLCKTTDTVRGIALALRTAAR
Mb1528c|M.bovis_AF2122/97 -------------------------MTAPAICNTTETVHGIATSLGAVAR
Rv1491c|M.tuberculosis_H37Rv -------------------------MTAPAICNTTETVHGIATSLGAVAR
MAV_3279|M.avium_104 -------------------------MTGSATSKIAETLRDLGCAIGAAAR
MAB_2713|M.abscessus_ATCC_1997 -----------------------------------MGFRQIGRTAVATLR
Mflv_3651|M.gilvum_PYR-GCK ----------------------MRAVPLVTVKSVVSTVRDAASAVAATLA
Mvan_2761|M.vanbaalenii_PYR-1 MMAVTPLSAGGRLNGVTIQPVRHGPVPLVIVKSVVSALRTVGAAVVATLK
MSMEG_3157|M.smegmatis_MC2_155 ------------------------------MKAVVSTLRSIRAAVLTTAA
TH_0612|M.thermoresistible__bu ------------------------------VNTVVNTLRAVRAAVVTTTA
.: : :.
MUL_1504|M.ulcerans_Agy99 QMSRLRKVSTVAGITLLVAVALLVPLPTAVQMRDWASSVGPWLPLVFLLV
MMAR_2301|M.marinum_M QMSRLRKVSTVAGITLLVAVALLVPLPTAVQMRDWASSVGPWLPLVFLLV
Mb1528c|M.bovis_AF2122/97 QASLPRIVGTVVGITVLVVVALLVPVPTAVELRDWAKSLGAWFPLAFLLV
Rv1491c|M.tuberculosis_H37Rv QASLPRIVGTVVGITVLVVVALLVPVPTAVELRDWAKSLGAWFPLAFLLV
MAV_3279|M.avium_104 GVSRSRIAWTVAGITALVVLASLIPLPSPVQMRDWAQSVGPWFPLAFLLA
MAB_2713|M.abscessus_ATCC_1997 RLPRRQLLWTGAALLIVIAAVRFVPLPTALQMRDWARDLGPWFPLAFLGA
Mflv_3651|M.gilvum_PYR-GCK TLPRTRLLAMAATIVILVAVAYFVPLPTALQVRDWATSAGPWFPLVFFVA
Mvan_2761|M.vanbaalenii_PYR-1 TAPRGRLAATLTAIVILVAVALLVPLPTALQLRDWATSVGSWFPLAFFAA
MSMEG_3157|M.smegmatis_MC2_155 TLSTRRIVAILAGIVILVAVALLVPLPTAMQLRDWATSVGPWFPLAFLGA
TH_0612|M.thermoresistible__bu QVPRHRLAMLAGGIVMLAAVVLLVPVPTAVQLRDWATSAGPWFPLAFLAA
. : : : . . ::*:*:.:::**** . *.*:**.*: .
MUL_1504|M.ulcerans_Agy99 HIVVTVPPFPRTAFTVAGGLLFGPVLGIVIAVTASTASAVIALLLVRAAG
MMAR_2301|M.marinum_M HIVVTVPPFPRTAFTVAGGLLFGPVLGIVIAVTASTASAVIALLLVRAAG
Mb1528c|M.bovis_AF2122/97 HTVVTVPPFPRTAFTLAAGLLFGSVVGVFIAVVGSTASAVIAMLLVRATG
Rv1491c|M.tuberculosis_H37Rv HTVVTVPPFPRTAFTLAAGLLFGSVVGVFIAVVGSTASAVIAMLLVRATG
MAV_3279|M.avium_104 HIVVTVVPVPRTAFTLAAGLLFGPLLGVAIAVAASTASAMIAMLLVRAAG
MAB_2713|M.abscessus_ATCC_1997 HTIVTVLPFPRTAFTLAAGLLFGTQLGVLLAVVASTASAVLALWAVRALG
Mflv_3651|M.gilvum_PYR-GCK HVVVTVFPFPRTAFTLAAGLLFGPALGVPIAVAASTLSAVIALLLVRTAG
Mvan_2761|M.vanbaalenii_PYR-1 HVVVTVFPFPRTAFTLAAGLLFGPVLGIPIAVAASTVSAVVALLLVRVAG
MSMEG_3157|M.smegmatis_MC2_155 HILVTVFPFPRTAFTLAAGLLFGPALGIAIAVSASAISAVLALLLIRAAG
TH_0612|M.thermoresistible__bu HVVVTVLPFPRTAFTLAAGLLFGPLLGVLLAVLASTISAVVTLIGVRALG
* :*** *.******:*.*****. :*: :** .*: **:::: :*. *
MUL_1504|M.ulcerans_Agy99 WRLNRLVPHGAVDRLDERLRERGWLAILSLRLIPAVPFAALNYGAGASAV
MMAR_2301|M.marinum_M WRLNRLVPHGAVDRLDERLRERGWLAILSLRLIPAVPFAALNYGAGASAV
Mb1528c|M.bovis_AF2122/97 WQLNSLVRRRAINRLDERLRERGWLAILSLRLIPVVPFAAINYAAGASGV
Rv1491c|M.tuberculosis_H37Rv WQLNSLVRRRAINRLDERLRERGWLAILSLRLIPVVPFAAINYAAGASGV
MAV_3279|M.avium_104 WRLTRLVRHRSMDTVEERLRQRGWLAIVSLRLIPAVPFSALNYAAGASSV
MAB_2713|M.abscessus_ATCC_1997 WKLSALHHRPAVKSVDDQLRRRGWIAVMSMRLIPAVPFSVLNYAAGASAV
Mflv_3651|M.gilvum_PYR-GCK WQVHKLVEHPRVDAIDQRLRERGWPVVLSARLIFALPFSVLNYAAGASSV
Mvan_2761|M.vanbaalenii_PYR-1 WQVSRLVVHPRVDAVDKRLRERGWPVVLATRMIPAVPFSVLNYAAGASSV
MSMEG_3157|M.smegmatis_MC2_155 WQLSRLVSHPRIDKLDARLRQRGWPVILSTRLIPAVPFSVLNYAAGASAV
TH_0612|M.thermoresistible__bu LRWTRLIAHPRIAAIDDRLRQRGWPAVISMRLIPAVPFSILNYAAGASAV
: * : : :: :**.*** .::: *:* .:**: :**.****.*
MUL_1504|M.ulcerans_Agy99 RVVPYTLSTLAGLLPGTVAVVILGSALAGDGSPLLVLVSVCTAALGMSGL
MMAR_2301|M.marinum_M RVVPYTLSTLAGLLPGTVAVVILGSALAGDGSPLLVLVSVCTAALGMSGL
Mb1528c|M.bovis_AF2122/97 RILSFAWATLAGLLPGTAAVVILGDAFAGSGSPLLILVSVCTGALGLTGL
Rv1491c|M.tuberculosis_H37Rv RILSFAWATLAGLLPGTAAVVILGDAFAGSGSPLLILVSVCTGALGLTGL
MAV_3279|M.avium_104 RVLPYGLATLAGLLPGTAAVVILGDALAGHPSSLLYLVSALTSALGLTGL
MAB_2713|M.abscessus_ATCC_1997 RALPYTLATFVGLLPGTLAVVVLGDALTGHISPTLFAVSLATSAIGVLGI
Mflv_3651|M.gilvum_PYR-GCK RVLPYTLATLFGVLPGTAAVVILGDALTGRVSPMLLVVSLCTALVGLAVL
Mvan_2761|M.vanbaalenii_PYR-1 RLMPYTLATVVGVLPGTSAIVILGDALTGRVSPLLVLVSLCTAAVGVAGL
MSMEG_3157|M.smegmatis_MC2_155 RLVPYALATLVGLLPGTAAVVILGDALTGNVSPLLFVVSACTAALGLAGL
TH_0612|M.thermoresistible__bu RVGPYTLATLVGLLPXXXSGRRTTAGSPDGRHPAVPLSGPSYGPLSGPGR
* .: :*. *:** : . .. . : . . :.
MUL_1504|M.ulcerans_Agy99 VFEVRQYRRHHRHQAPSDHG-PVLEPACEPLGV
MMAR_2301|M.marinum_M VFEVRQYRRHHRHQAPSDHG-PVLEPACEPLGV
Mb1528c|M.bovis_AF2122/97 VYEIRNYRRQHR-RMPGYDD-PVREPALI----
Rv1491c|M.tuberculosis_H37Rv VYEIRNYRRQHR-RMPGYDD-PVREPALI----
MAV_3279|M.avium_104 IIEIRHFRRHHRRAHRHRDDEPSPEPATIG---
MAB_2713|M.abscessus_ATCC_1997 LYEIRRYKRTHT-EIPDPESSGAPERVGR----
Mflv_3651|M.gilvum_PYR-GCK ALEIRSHRR-----AGAADARP-----------
Mvan_2761|M.vanbaalenii_PYR-1 IYEIRTHRR-----SQTPVTTV-----------
MSMEG_3157|M.smegmatis_MC2_155 VFEIRSHRREQRAAAGAADTAASNL--------
TH_0612|M.thermoresistible__bu SRGPRARRPRPRR--------------------
* :