For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PSHPNQVDVRGPRFAAWITTMVLILTLVVSISSTRAAAVILGVQTMVFAVTALRGPRRSPYGFLFAKLAA ARLGPARELEPVPPLRFAQLVGLVFAAAGVVGFALGVPLWGVVATSLALFAAFLNAAFGICLGCQLYPLV ARLRRAPIA*
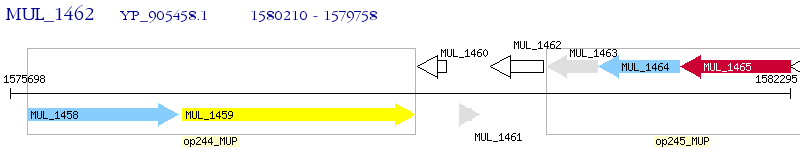
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1462 | - | - | 100% (150) | hypothetical protein MUL_1462 |
| M. ulcerans Agy99 | MUL_0433 | - | 3e-49 | 68.09% (141) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1647 | - | 3e-50 | 65.97% (144) | hypothetical protein Mflv_1647 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02197 | - | 1e-46 | 64.03% (139) | integral membrane protein |
| M. abscessus ATCC 19977 | MAB_0741c | - | 2e-51 | 68.79% (141) | hypothetical protein MAB_0741c |
| M. marinum M | MMAR_3648 | - | 2e-80 | 99.33% (150) | hypothetical protein MMAR_3648 |
| M. avium 104 | MAV_0757 | - | 3e-52 | 68.06% (144) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_5788 | - | 1e-51 | 70.21% (141) | integral membrane protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5100 | - | 4e-48 | 64.79% (142) | hypothetical protein Mvan_5100 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1647|M.gilvum_PYR-GCK MSNITAADSP---AQVDVRGPRFVAWVTTAVLVAALLISVQNAALAAALL
Mvan_5100|M.vanbaalenii_PYR-1 MSNITQSSAP---TQIDVRGPRFVAWVTTVVLVATLLVSAQHPAAAAVLL
MUL_1462|M.ulcerans_Agy99 MPSHP--------NQVDVRGPRFAAWITTMVLILTLVVSISSTRAAAVIL
MMAR_3648|M.marinum_M MPSHP--------SQVDVRGPRFAAWITTMVLILTLVVSISSTRAAAVIL
MAB_0741c|M.abscessus_ATCC_199 MTTANTTPA---ISQVDVRGPRFSAWVTTTVLIITLAVSAVSPTAAAVIL
MSMEG_5788|M.smegmatis_MC2_155 MSSSITDTARPRLDQVDVRGPRFAAWVTTAVLIVALLVSGVSSAAAAVVL
MLBr_02197|M.leprae_Br4923 MTINNTTTGV---EVVDVRGPRFAAWVTAAVLQTVLIISVVSPLAAALLL
MAV_0757|M.avium_104 MPSSNTTTQP---DLVDVRGPRFAAWVTTAVLVLALAVSAVSPPAAAVIL
*. :******* **:*: ** .* :* . ** :*
Mflv_1647|M.gilvum_PYR-GCK GIQTLVFAVGAVAGPRRHPYGVAFAALIAPRLGPAREREPVAPLKFAQLV
Mvan_5100|M.vanbaalenii_PYR-1 GAQAVVFAIGAVSGPRRHPYGRVFAALIAPRLAPATDREPVAPLKFAQLV
MUL_1462|M.ulcerans_Agy99 GVQTMVFAVTALRGPRRSPYGFLFAKLAAARLGPARELEPVPPLRFAQLV
MMAR_3648|M.marinum_M GVQTMVFAVTALRGPRRSPYGFLFAKLAAARLGPARELEPVPPLRFAQLV
MAB_0741c|M.abscessus_ATCC_199 GLQAVVFAIGALLGPHRAPYGTIFRTLIQPRLSPVTEREPVPPLQFAQLL
MSMEG_5788|M.smegmatis_MC2_155 AAQALVFAIGALGGPRRQPYGRLFATLVAPRLGPVTEREPVPPLKFAQLV
MLBr_02197|M.leprae_Br4923 VLQAVVFATGTLGGPRRHPYGRVFAALVAPRLGPVKEREPVPPLKFAQLL
MAV_0757|M.avium_104 AVQAVVFAIGAVGGPRKHPYGRVFAAVVAPRLGPVREREPIPPLKFAQLV
*::*** :: **:: *** * : .**.*. : **:.**:****:
Mflv_1647|M.gilvum_PYR-GCK GFVFAALSTLGFAGGVMPLGLIATGLALAAALLNAAFGICLGCQLYPLIA
Mvan_5100|M.vanbaalenii_PYR-1 GFAFAVVGVLGFASGLAPLGLVATGFALVAALLNAAFGICLGCQLYPLVT
MUL_1462|M.ulcerans_Agy99 GLVFAAAGVVGFALGVPLWGVVATSLALFAAFLNAAFGICLGCQLYPLVA
MMAR_3648|M.marinum_M GLVFAAAGVVGFALGVPLWGVVATSLALFAAFLNAAFGICLGCQLYPLVA
MAB_0741c|M.abscessus_ATCC_199 GFTFAAVGTVGFATGVTLLGVIATALALGAAFLNAAFGICLGCQIYPLIT
MSMEG_5788|M.smegmatis_MC2_155 GFVFAAVGAVGFAFGISAIGLAATAFALVAAFLNAAFDICLGCQIYPLVA
MLBr_02197|M.leprae_Br4923 GFVFVVCGVTGFAAGAFLVGVIATAAALVAAFLNAAFGICLGCQLYPVVV
MAV_0757|M.avium_104 GLIFAVLGAAGFAAGASLFGLVATAAALAAAFLNAAFGICLGCQLYPLVA
*: *.. .. *** * *: **. ** **:*****.******:**::.
Mflv_1647|M.gilvum_PYR-GCK RLTPTP--------
Mvan_5100|M.vanbaalenii_PYR-1 RLNPTR--------
MUL_1462|M.ulcerans_Agy99 RLRR-APIA-----
MMAR_3648|M.marinum_M RLRR-APIA-----
MAB_0741c|M.abscessus_ATCC_199 RLRRTQPSAQ----
MSMEG_5788|M.smegmatis_MC2_155 RLRRTSEYA-----
MLBr_02197|M.leprae_Br4923 WFRRTAAPRNRNPG
MAV_0757|M.avium_104 RFRRPARST-----
: