For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSSITDTARPRLDQVDVRGPRFAAWVTTAVLIVALLVSGVSSAAAAVVLAAQALVFAIGALGGPRRQPY GRLFATLVAPRLGPVTEREPVPPLKFAQLVGFVFAAVGAVGFAFGISAIGLAATAFALVAAFLNAAFDIC LGCQIYPLVARLRRTSEYA
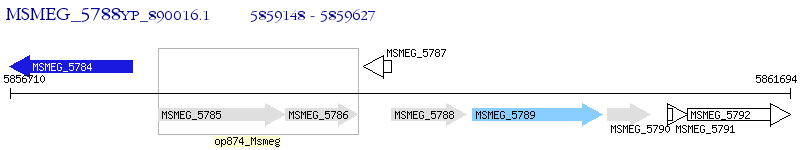
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5788 | - | - | 100% (159) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1647 | - | 5e-52 | 66.01% (153) | hypothetical protein Mflv_1647 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02197 | - | 2e-53 | 69.44% (144) | integral membrane protein |
| M. abscessus ATCC 19977 | MAB_0741c | - | 2e-57 | 69.48% (154) | hypothetical protein MAB_0741c |
| M. marinum M | MMAR_4867 | - | 4e-54 | 68.83% (154) | integral membrane protein |
| M. avium 104 | MAV_0757 | - | 1e-58 | 73.51% (151) | integral membrane protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0433 | - | 6e-52 | 66.88% (154) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_5100 | - | 1e-55 | 68.63% (153) | hypothetical protein Mvan_5100 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5788|M.smegmatis_MC2_155 MSSSITDTARPRLDQVDVRGPRFAAWVTTAVLIVALLVSGVSSAAAAVVL
MAB_0741c|M.abscessus_ATCC_199 MT---TANTTPAISQVDVRGPRFSAWVTTTVLIITLAVSAVSPTAAAVIL
MLBr_02197|M.leprae_Br4923 MT---INNTTTGVEVVDVRGPRFAAWVTAAVLQTVLIISVVSPLAAALLL
MAV_0757|M.avium_104 MP---SSNTTTQPDLVDVRGPRFAAWVTTAVLVLALAVSAVSPPAAAVIL
MMAR_4867|M.marinum_M MS---STNTRTAVELVDVRGPRFAAWVTTAVLLVVLIISALNSAAAAVVL
MUL_0433|M.ulcerans_Agy99 MS---STITRTAVELVDVRGPRFAAWVTTAVLLVVLIISALN-PAAAVVL
Mflv_1647|M.gilvum_PYR-GCK MS---NITAADSPAQVDVRGPRFVAWVTTAVLVAALLISVQNAALAAALL
Mvan_5100|M.vanbaalenii_PYR-1 MS---NITQSSAPTQIDVRGPRFVAWVTTVVLVATLLVSAQHPAAAAVLL
*. :******* ****:.** .* :* ** :*
MSMEG_5788|M.smegmatis_MC2_155 AAQALVFAIGALGGPRRQPYGRLFATLVAPRLGPVTEREPVPPLKFAQLV
MAB_0741c|M.abscessus_ATCC_199 GLQAVVFAIGALLGPHRAPYGTIFRTLIQPRLSPVTEREPVPPLQFAQLL
MLBr_02197|M.leprae_Br4923 VLQAVVFATGTLGGPRRHPYGRVFAALVAPRLGPVKEREPVPPLKFAQLL
MAV_0757|M.avium_104 AVQAVVFAIGAVGGPRKHPYGRVFAAVVAPRLGPVREREPIPPLKFAQLV
MMAR_4867|M.marinum_M GLQATAFAIGAFAGPRRHPYGVLFSTFVAPRIAAVREREPVPPLKFAQLV
MUL_0433|M.ulcerans_Agy99 GLQATAFAVGAFAGPRRHPYGVLFSAFVAPRIAAVREREPVPPLKFAQLV
Mflv_1647|M.gilvum_PYR-GCK GIQTLVFAVGAVAGPRRHPYGVAFAALIAPRLGPAREREPVAPLKFAQLV
Mvan_5100|M.vanbaalenii_PYR-1 GAQAVVFAIGAVSGPRRHPYGRVFAALIAPRLAPATDREPVAPLKFAQLV
*: .** *:. **:: *** * :.: **:... :***:.**:****:
MSMEG_5788|M.smegmatis_MC2_155 GFVFAAVGAVGFAFGISAIGLAATAFALVAAFLNAAFDICLGCQIYPLVA
MAB_0741c|M.abscessus_ATCC_199 GFTFAAVGTVGFATGVTLLGVIATALALGAAFLNAAFGICLGCQIYPLIT
MLBr_02197|M.leprae_Br4923 GFVFVVCGVTGFAAGAFLVGVIATAAALVAAFLNAAFGICLGCQLYPVVV
MAV_0757|M.avium_104 GLIFAVLGAAGFAAGASLFGLVATAAALAAAFLNAAFGICLGCQLYPLVA
MMAR_4867|M.marinum_M GLVFAVCGVAGFALGPFLVGAVATALALVAAFLNAAFGICLGCELYPLAA
MUL_0433|M.ulcerans_Agy99 GLVFAVCGVAGFALGPFLVGAVATALALVAAFLNAAFGICLGCELYPLAA
Mflv_1647|M.gilvum_PYR-GCK GFVFAALSTLGFAGGVMPLGLIATGLALAAALLNAAFGICLGCQLYPLIA
Mvan_5100|M.vanbaalenii_PYR-1 GFAFAVVGVLGFASGLAPLGLVATGFALVAALLNAAFGICLGCQLYPLVT
*: *.. .. *** * .* **. ** **:*****.*****::**: .
MSMEG_5788|M.smegmatis_MC2_155 RLRRTSEYA-----
MAB_0741c|M.abscessus_ATCC_199 RLRRTQPSAQ----
MLBr_02197|M.leprae_Br4923 WFRRTAAPRNRNPG
MAV_0757|M.avium_104 RFRRPARST-----
MMAR_4867|M.marinum_M RLRRDPHPA-----
MUL_0433|M.ulcerans_Agy99 RLRRDPHPA-----
Mflv_1647|M.gilvum_PYR-GCK RLTPTP--------
Mvan_5100|M.vanbaalenii_PYR-1 RLNPTR--------
: