For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VHTGYMGIREGGPYFDDLSVGQVFDWAPALTLSAGLAAAHQAILGDRLRLALDSSLCGAVTGAPAALAHP GLVCNAAIGQSTLVTQRVKANLFYRGLTFHRYPLIGDTIYTRTEVVALRANTAKPRRAPTGMAALRMTTI DQADRLVLDFYRCAMLPASADWDPAGAADDDLSGIGADAKPPAIDPTAHWDAEAYRTRVPGPHFDAGIAG KVMHSTGDLVSSAPELARLTLNIAATHHDSRVAGQRLVYGGHTIGLALAQASRLMPNLATVLGWSHCDHT GPVREGDTLYSELHVESAEPTANGGVLGLRSLVYAVGDSEPDRQVLDWRFTALQY
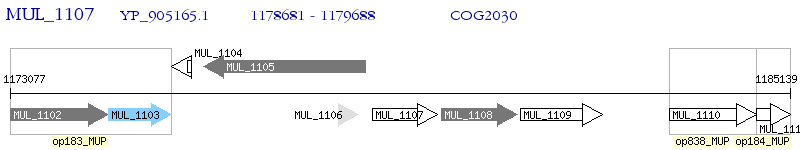
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1107 | - | - | 100% (335) | hypothetical protein MUL_1107 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0222 | - | 1e-155 | 78.64% (337) | hypothetical protein Mb0222 |
| M. gilvum PYR-GCK | Mflv_0445 | - | 1e-133 | 71.69% (332) | hypothetical protein Mflv_0445 |
| M. tuberculosis H37Rv | Rv0216 | - | 1e-155 | 78.64% (337) | hypothetical protein Rv0216 |
| M. leprae Br4923 | MLBr_02627 | - | 1e-145 | 75.52% (335) | hypothetical protein MLBr_02627 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0456 | - | 0.0 | 99.10% (335) | hypothetical protein MMAR_0456 |
| M. avium 104 | MAV_4957 | - | 1e-154 | 78.87% (336) | p40 protein |
| M. smegmatis MC2 155 | MSMEG_0261 | - | 1e-130 | 70.66% (334) | p40 protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0222 | - | 1e-132 | 72.95% (329) | hypothetical protein Mvan_0222 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_1107|M.ulcerans_Agy99 -MHTGYMGIREGGPYFDDLSVGQVFDWAPALTLSAGLAAAHQAILGDRLR
MMAR_0456|M.marinum_M -MHTGYMGIREGGPYFDDLSVGQVFDWAPALTLSAGLAAAHQAILGDRLR
Mb0222|M.bovis_AF2122/97 -MASGYGGIRVGGPYFDDLSKGQVFDWAPGVTLSLGLAAAHQSIVGNRLR
Rv0216|M.tuberculosis_H37Rv -MASGYGGIRVGGPYFDDLSKGQVFDWAPGVTLSLGLAAAHQSIVGNRLR
MAV_4957|M.avium_104 MTSDGYARIREGGPYFDDLSVGQVFDWAPSVTLSSGLAAAHQAILGDRLR
MLBr_02627|M.leprae_Br4923 MTANGHAGRREGGPYFDDLSIGQVFDWAPAVTLTSGMAAVHQAILGDRMR
Mflv_0445|M.gilvum_PYR-GCK MPAVTLYGQIPGGPFFDDLSVGQVFDAAPSMTLTPGVAATHQAILGDRLR
Mvan_0222|M.vanbaalenii_PYR-1 ---------MSGGPHFDDLRVGQVFDSAPSMTLTAGVAATHQAIIGDRMR
MSMEG_0261|M.smegmatis_MC2_155 ---MTAHGRRSGGPYFDDLAVGQVFDTAPSMTLTEGAAAAHQAVIGDRLR
***.**** ***** **.:**: * **.**:::*:*:*
MUL_1107|M.ulcerans_Agy99 LALDSSLCGAVTGAPAALAHPGLVCNAAIGQSTLVTQRVKANLFYRGLTF
MMAR_0456|M.marinum_M LALDSSLCGAVTGAPAALAHPGLVCNAAIGQSTLVTQRVKANLFYRGLTF
Mb0222|M.bovis_AF2122/97 LALDSDLCAAVTGMPGPLAHPGLVCDVAIGQSTLATQRVKANLFYRGLRF
Rv0216|M.tuberculosis_H37Rv LALDSDLCAAVTGMPGPLAHPGLVCDVAIGQSTLATQRVKANLFYRGLRF
MAV_4957|M.avium_104 LALDADLCAAVTGLPAPLAHPGLVCDVAIGQSTLVTQRVKANLFYRGLVF
MLBr_02627|M.leprae_Br4923 LALDAELSTTVIGTHAMLAHPGLVCDVAIGQSTLVTQRVKANLFYRGLTF
Mflv_0445|M.gilvum_PYR-GCK LPLDADLSSAVTGSTAALAHPALVCDVAIGQSTLVTQRVKANLFYRGLTF
Mvan_0222|M.vanbaalenii_PYR-1 LPLDGELSLSVAGATAPLAHPALVCDVAIGQSTLATQRVKANLFYRGLTF
MSMEG_0261|M.smegmatis_MC2_155 LSLDAELSSAVTGEPGPLAHPALVCDVAIGQSTLVTQRVKANLFYRGLAF
*.**..*. :* * . ****.***:.*******.************* *
MUL_1107|M.ulcerans_Agy99 HRYPLIGDTIYTRTEVVALRANTAKPRRAPTGMAALRMTTIDQADRLVLD
MMAR_0456|M.marinum_M HRYPLIGDTIYTRTEVVALRANTAKPGRAPTGMAALRMTTIDQADRLVLD
Mb0222|M.bovis_AF2122/97 HRFPAVGDTLYTRTEVVGLRANSPKPGRAPTGLAGLRMTTIDRTDRLVLD
Rv0216|M.tuberculosis_H37Rv HRFPAVGDTLYTRTEVVGLRANSPKPGRAPTGLAGLRMTTIDRTDRLVLD
MAV_4957|M.avium_104 HRFPVIGDSLYTRTEVVGLRANSPKPGRAPTGMAALRMITIDQADQLVLD
MLBr_02627|M.leprae_Br4923 HRFPVIGDTLYTSTEVVGLQANSAKPGRPPTGMAALRITTTDQHDRLVLD
Mflv_0445|M.gilvum_PYR-GCK HRYPVLGDTIYTRTEVVGLKQNSAKPGRAATGLAALRMTTIDGIGRLVLD
Mvan_0222|M.vanbaalenii_PYR-1 YRYPVIGDSLFTRTEVVGLRQNTAKPGRAPTGLAALRMTTIDGVGRLILD
MSMEG_0261|M.smegmatis_MC2_155 HRFPVIGDTLTTRTEVVGLKQNSNKPGRAPTGLAALRMTTTDHKDRLVLD
:*:* :**:: * ****.*: *: ** *..**:*.**: * * .:*:**
MUL_1107|M.ulcerans_Agy99 FYRCAMLPASADWDP--AGAADDDLSGIGADAKPPAI-----DPTAHWDA
MMAR_0456|M.marinum_M FYRCAMLPASADWDP--AGAADDDLSGIGADAKPPAI-----DPTAHWDA
Mb0222|M.bovis_AF2122/97 FYRCAMLPASPDWKP--GAVPGDDLSRIGADAPAPAA-----DPTAHWDG
Rv0216|M.tuberculosis_H37Rv FYRCAMLPASPDWKP--GAVPGDDLSRIGADAPAPAA-----DPTAHWDG
MAV_4957|M.avium_104 FYRCGMLPASPEFTPEAGSVPHDDLSGIGADVPGPAH-----DPTAQWDG
MLBr_02627|M.leprae_Br4923 FYRCAMLPASAAWHPHDALNRNDDLANVGADAVASAS-----DPTGQWDA
Mflv_0445|M.gilvum_PYR-GCK FYRCAMLPLSSDDVQ---TGHDDDLSTVGAGGSDAASGTRLVDPTRDWDD
Mvan_0222|M.vanbaalenii_PYR-1 FYRCAMLPLSDDTAQ---TGHDDDLSAIGAG---AAP---VADPTADWDA
MSMEG_0261|M.smegmatis_MC2_155 FYRCAMLPLSENGGD---TGYSDDLSAIGAD---ASP-----TPTAHWDA
****.*** * ***: :**. .: ** .**
MUL_1107|M.ulcerans_Agy99 EAYRTRVPGPHFD---AGIAGKVMHSTGDLVSSAPELARLTLNIAATHHD
MMAR_0456|M.marinum_M EAYRTRVPGPHFD---AGIAGKVMHSTGDLVSSAPELARLTLNIAATHHD
Mb0222|M.bovis_AF2122/97 AVFRKRVPGPHFD---AGIAGAVLHSTADLVSGAPELARLTLNIAATHHD
Rv0216|M.tuberculosis_H37Rv AVFRKRVPGPHFD---AGIAGAVLHSTADLVSGAPELARLTLNIAATHHD
MAV_4957|M.avium_104 DVFRTEVPGPHFD---PALAGSVLHSTGDVVSSAPELARLTLNIAATHHD
MLBr_02627|M.leprae_Br4923 AAFRERVPGPHFD---AGITGAVLRSTGDIVSSAPDLARLTLNIASTHHD
Mflv_0445|M.gilvum_PYR-GCK AAFRDRVPGPHFD---PALAGTVLRSTADVVSSAPELARLSLNIAATHHD
Mvan_0222|M.vanbaalenii_PYR-1 EAFRTRVPGPHFD---PALVGTVGRSTADVVSSAPELARLSLNIAATHHD
MSMEG_0261|M.smegmatis_MC2_155 DAYRARVPGPHLDDLRDELAGAVLRSTADVVSSAPELARLTLNIAATHHD
.:* .*****:* :.* * :**.*:**.**:****:****:****
MUL_1107|M.ulcerans_Agy99 SRVAGQRLVYGGHTIGLALAQASRLMPNLATVLGWSHCDHTGPVREGDTL
MMAR_0456|M.marinum_M SRVAGQRLVYGGHTIGLALAQASRLMPNLVTVLGWEHCDHTGPVREGDTL
Mb0222|M.bovis_AF2122/97 WRVSGRRLVYGGHTIGLALAQATRLLPNLATVLDWESCDHTAPVHEGDTL
Rv0216|M.tuberculosis_H37Rv WRVSGRRLVYGGHTIGLALAQATRLLPNLATVLDWESCDHTAPVHEGDTL
MAV_4957|M.avium_104 SRVGGRRLVYGGHTIGLALAQAARLLPNLVTVLGWRSCDHTGPVHEGDTL
MLBr_02627|M.leprae_Br4923 SRVRGLRLVYGGHTIGLALAQAGRMLPNLATVLNWRSCDHTGPVHEGDTL
Mflv_0445|M.gilvum_PYR-GCK SRVAGKRLVYGGHTIGLALAQASRLLPNLVTVLGWESCDHTGPVHEGDTL
Mvan_0222|M.vanbaalenii_PYR-1 SRVAGRRLVYGGHTIGLALAQATRLLPNLVTVLGWESCDHTGPVYENDTL
MSMEG_0261|M.smegmatis_MC2_155 WRSGGRRLVYGGHTIGLALAQATRLLPNLVTVLGWESCDHTGPVHEGDTV
* * **************** *::***.***.* ****.** *.**:
MUL_1107|M.ulcerans_Agy99 YSELHVESAEPTANG--GVLGLRSLVYAVGDS--EPDRQVLDWRFTALQY
MMAR_0456|M.marinum_M YSELHVESAEPTANG--GVLGLRSLVYAVGDS--EPDRQVLDWRFTALQY
Mb0222|M.bovis_AF2122/97 YSELHIESAQAHADG--GVLGLRSLVYAVSDSASEPDRQVLDWRFSALQF
Rv0216|M.tuberculosis_H37Rv YSELHIESAQAHADG--GVLGLRSLVYAVSDSASEPDRQVLDWRFSALQF
MAV_4957|M.avium_104 YSELHIESARPTEHG--GVLGLRSLVYAVSDTPDERDRAVLDWRFDALQF
MLBr_02627|M.leprae_Br4923 YSELHVESAEATEQG--GVLGLRSLVYAVSAAG-GSDHLVLDWRFTVLQF
Mflv_0445|M.gilvum_PYR-GCK YSELRVESVQPRPDGRGGTVALRSTVFAAS-DET-EDRPVLDWRFTGLLF
Mvan_0222|M.vanbaalenii_PYR-1 YSDLTVESADPLPDGRGAVLRLRSVVHAAG-DEPGADRQVLEWRFSALHF
MSMEG_0261|M.smegmatis_MC2_155 FSELHVESIEAN------VVGLRSIVFAAGGPDGGADRQVLDWRFSALLF
:*:* :** . .: *** *.*.. *: **:*** * :