For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLFRNVAIVAHVDHGKTTLVDAMLRQSGALTERGEVQERVMDSGDLEREKGITILAKNTAVHRHHPDGTV TVINVIDTPGHADFGGEVERGLSMVDGVLLLVDASEGPLPQTGFVLRKALSAHLPVILVVNKTDRPDARI KEVVEASHDLLLDVASDLDDEAAAAAEHALGLPTLYASGRAGIASTIEPADGQAPDGTNLDPLFDVLLEH VPPPQGDSEAPLQALVTNLDASAFLGRLALVRIYNGKLRKGQQVAWLREVDGVPVVTSAKITELLATEGV ERSTTDEAVAGDIVAVAGLPEIMIGDTLADPDHAHALPRITVDEPAISVTVGTNTSPLAGKVSGHKLTAR MVRGRLDTELIGNVSIRVVDIGRPDAWEVQGRGELALAVLVETMRREGFELTVGKPQVVTKTIDGQLHEP FEAMTIDCPDEFVGAITQLMAGRKGRVEEMTNHAAGWVRMDFIVPSRGLIGFRTDFLTLTRGTGIANAVF DGYRPWAGEIRARHTGSLVSDRSGVITPFALLQLADRGQFFVEPGEDTYEGMVVGINPRAEDLDINVTRE KKLTNMRSSTADVIETLAKPLELDLERAIEFCAPDECVEVTPEAIRVRKVELDATSRARSRSRAKARG
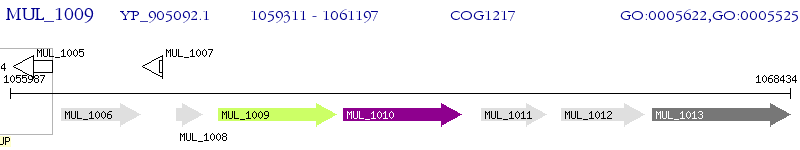
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1009 | typA | - | 100% (628) | GTP-binding translation elongation factor TypA |
| M. ulcerans Agy99 | MUL_0765 | fusA1 | 1e-35 | 27.98% (511) | elongation factor G |
| M. ulcerans Agy99 | MUL_3667 | lepA | 6e-28 | 27.33% (494) | GTP-binding protein LepA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1197 | typA | 0.0 | 86.94% (628) | GTP-binding translation elongation factor TypA |
| M. gilvum PYR-GCK | Mflv_2163 | - | 0.0 | 88.36% (627) | GTP-binding protein TypA |
| M. tuberculosis H37Rv | Rv1165 | typA | 0.0 | 86.94% (628) | GTP-binding translation elongation factor TypA |
| M. leprae Br4923 | MLBr_01498 | - | 0.0 | 89.33% (628) | putative GTP-binding, protein elongation factor |
| M. abscessus ATCC 19977 | MAB_1310 | - | 0.0 | 87.84% (625) | GTP-binding translation elongation factor |
| M. marinum M | MMAR_4289 | typA | 0.0 | 99.36% (628) | GTP-binding translation elongation factor TypA |
| M. avium 104 | MAV_1307 | typA | 0.0 | 91.10% (629) | GTP-binding protein TypA/BipA |
| M. smegmatis MC2 155 | MSMEG_5132 | typA | 0.0 | 89.15% (627) | GTP-binding protein TypA/BipA |
| M. thermoresistible (build 8) | TH_1877 | typA | 0.0 | 85.35% (628) | GTP-binding protein TypA/BipA |
| M. vanbaalenii PYR-1 | Mvan_4540 | - | 0.0 | 88.14% (624) | GTP-binding protein TypA |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1197|M.bovis_AF2122/97 -------------------------MPFRNVAIVAHVDHGKTTLVDAMLR
Rv1165|M.tuberculosis_H37Rv -------------------------MPFRNVAIVAHVDHGKTTLVDAMLR
MUL_1009|M.ulcerans_Agy99 -------------------------MLFRNVAIVAHVDHGKTTLVDAMLR
MMAR_4289|M.marinum_M -------------------------MLFRNVAIVAHVDHGKTTLVDAMLR
MLBr_01498|M.leprae_Br4923 -------------------------MPFRNVAIVAHVDHGKTTLVDAMLR
Mflv_2163|M.gilvum_PYR-GCK ------------MQCTATITDVNARPDFRNVAIVAHVDHGKTTLVDAMLR
Mvan_4540|M.vanbaalenii_PYR-1 ---------------------MNARPDFRNVAIVAHVDHGKTTLVDAMLR
MSMEG_5132|M.smegmatis_MC2_155 ---------------------MSSHPDFRNVAIVAHVDHGKTTLVDAMLR
MAB_1310|M.abscessus_ATCC_1997 MSGVTPIVRRFRELWAVLPELADVTHSFRNVAIVAHVDHGKTTLVDAMLK
MAV_1307|M.avium_104 -------------------------MPFRNVAIVAHVDHGKTTLVDAMLR
TH_1877|M.thermoresistible__bu -------------------------VNFRNVAIVAHVDHGKTTLVDAMLR
**********************:
Mb1197|M.bovis_AF2122/97 QSGALRERGE-LQERVMDTGDLEREKGITILAKNTAVHRHHPDGTVTVIN
Rv1165|M.tuberculosis_H37Rv QSGALRERGE-LQERVMDTGDLEREKGITILAKNTAVHRHHPDGTVTVIN
MUL_1009|M.ulcerans_Agy99 QSGALTERGE-VQERVMDSGDLEREKGITILAKNTAVHRHHPDGTVTVIN
MMAR_4289|M.marinum_M QSGALTERGE-VQERVMDSGDLEREKGITILAKNTAVHRHHPDGTVTVIN
MLBr_01498|M.leprae_Br4923 QSGALHERGQ-VQERVMDTGDLEREKGITILAKNTAVHCHNSDGTVTVIN
Mflv_2163|M.gilvum_PYR-GCK QSGALTHRGDDAIERLMDSGDLEKEKGITILAKNTAVHRTHPDGTVTVIN
Mvan_4540|M.vanbaalenii_PYR-1 QSGALSHRGDDAIERLMDSGDLEKEKGITILAKNTAVHRHHADGSMTVIN
MSMEG_5132|M.smegmatis_MC2_155 QSGALTHRGDDSTERIMDSGDLEREKGITILAKNTAVHRHNADGTVTVIN
MAB_1310|M.abscessus_ATCC_1997 QSGALSHRGDDAVERLMDSGDLEKEKGITILAKNTAVHRHHADGSMTVIN
MAV_1307|M.avium_104 QSGALHHRGDDTQERILDSGDLEKEKGITILAKNTAVHRHHPDGSVTVIN
TH_1877|M.thermoresistible__bu QSGALKARGDDTTERIMDSGDLEREKGITILAKNTAVHRHHPDGSVTVIN
***** **: **::*:****:************** :.**::****
Mb1197|M.bovis_AF2122/97 VIDTPGHADFGGEVERGLSMVDGVLLLVDASEGPLPQTRFVLRKALAAHL
Rv1165|M.tuberculosis_H37Rv VIDTPGHADFGGEVERGLSMVDGVLLLVDASEGPLPQTRFVLRKALAAHL
MUL_1009|M.ulcerans_Agy99 VIDTPGHADFGGEVERGLSMVDGVLLLVDASEGPLPQTGFVLRKALSAHL
MMAR_4289|M.marinum_M VIDTPGHADFGGEVERGLSMVDGVLLLVDASEGPLPQTRFVLRKALSAHL
MLBr_01498|M.leprae_Br4923 VIDTPGHADFGGEVERGLSMVDGVLLLVDASEGPLPQTRFVLRKTLAAHL
Mflv_2163|M.gilvum_PYR-GCK VIDTPGHADFGGEVERGLSMVDGVVLLVDASEGPLPQTRFVLRKALAAHL
Mvan_4540|M.vanbaalenii_PYR-1 VIDTPGHADFGGEVERGLSMVDGVVLLVDASEGPLPQTRFVLRKALAAHL
MSMEG_5132|M.smegmatis_MC2_155 VIDTPGHADFGGEVERGLSMVDGVLLLVDASEGPLPQTRFVLRKALAAHL
MAB_1310|M.abscessus_ATCC_1997 VIDTPGHADFGGEVERGLSMVDGVLLLVDASEGPLPQTRFVLRKALAAHL
MAV_1307|M.avium_104 VIDTPGHADFGGEVERGLSMVDGVLLLVDASEGPLPQTRFVLRKALAAHL
TH_1877|M.thermoresistible__bu VIDTPGHADFGGEVERGLSMVDGVLLLVDASEGPLPQTRFVLRKALAAHL
************************:************* *****:*:***
Mb1197|M.bovis_AF2122/97 PVILVVNKTDRPDARIAEVVDASHDLLLDVASDLDDEAAAAAEHALGLPT
Rv1165|M.tuberculosis_H37Rv PVILVVNKTDRPDARIAEVVDASHDLLLDVASDLDDEAAAAAEHALGLPT
MUL_1009|M.ulcerans_Agy99 PVILVVNKTDRPDARIKEVVEASHDLLLDVASDLDDEAAAAAEHALGLPT
MMAR_4289|M.marinum_M PVILVVNKTDRPDARIKEVVEASHDLLLDVASDLDDEAAAAAEHALGLPT
MLBr_01498|M.leprae_Br4923 PVILVVNKTDRPDARIAEVVEASHDLLLDVASDLDEEAAAAAERALGLPT
Mflv_2163|M.gilvum_PYR-GCK PVILVVNKTDRPDARIAEVVEESHDLLLDVASDLDEEAQAAAEKALDLPT
Mvan_4540|M.vanbaalenii_PYR-1 PVILVVNKTDRPDARIAEVVEESHDLLLDVASDLDEEAQTAAEKALDLPT
MSMEG_5132|M.smegmatis_MC2_155 PVILVVNKTDRPDARIAEVVSDSHDLLLDVASDLDEDAQAAAEKALDLPT
MAB_1310|M.abscessus_ATCC_1997 PVILVVNKTDRPDARIAEVVSDSHDLLLDVASDLDEEAQKAAEDALGLPT
MAV_1307|M.avium_104 PVILVVNKTDRPDARIAEVVEASHDLLLDVASDLDDEAAKAAEHALGLPT
TH_1877|M.thermoresistible__bu PVILVVNKTDRPDARIAEVVEASHDLLLDVAADLDEQAQQAAEHALGLPT
**************** ***. *********:***::* *** **.***
Mb1197|M.bovis_AF2122/97 LYASGRAGVASTTAPPDGQVPDGTNLDPLFEVLEKHVPPPKGEPDAPLQA
Rv1165|M.tuberculosis_H37Rv LYASGRAGVASTTAPPDGQVPDGTNLDPLFEVLEKHVPPPKGEPDAPLQA
MUL_1009|M.ulcerans_Agy99 LYASGRAGIASTIEPADGQAPDGTNLDPLFDVLLEHVPPPQGDSEAPLQA
MMAR_4289|M.marinum_M LYASGRAGIASTIEPADGQAPDGTNLDPLFDVLLEHVPPPQGDSEAPLQA
MLBr_01498|M.leprae_Br4923 LYASGRAGIASTEQPADGAVPTGDNLDPLFDVLMEHIPSPKGDPEAPLQA
Mflv_2163|M.gilvum_PYR-GCK LYASGRAGIASTTAPANGENPDGENLDPLFDVLLEHIPPPQGDPDAPLQA
Mvan_4540|M.vanbaalenii_PYR-1 LYASGRAGIASTVQPANGENPDGENLDPLFDVLLEHIPPPQGDPEAPLQA
MSMEG_5132|M.smegmatis_MC2_155 LYASGRAGVASTVQPADGEVPEGDNLDPLFDVLMKHIPPPSGDPEAPLQA
MAB_1310|M.abscessus_ATCC_1997 LYASGRAGVASTVEPANGEVPEGDNLDPLFDVLLEHIPPPKGDPEAPLQA
MAV_1307|M.avium_104 LYASGRAGVASTEQPADGEVPAGENLDPLFDVLLEHIPAPSGDPEAPLQA
TH_1877|M.thermoresistible__bu LYASGRAGIASTTPPPDGRVPDGTNLDPLFDVLLEHIPPPSGDPDQPLQA
********:*** *.:* * * ******:** :*:*.*.*:.: ****
Mb1197|M.bovis_AF2122/97 LVTNLDASTFLGRLALIRIYNGRIRKGQQVAWIRQVDGQQTVTTAKITEL
Rv1165|M.tuberculosis_H37Rv LVTNLDASTFLGRLALIRIYNGRIRKGQQVAWIRQVDGQQTVTTAKITEL
MUL_1009|M.ulcerans_Agy99 LVTNLDASAFLGRLALVRIYNGKLRKGQQVAWLREVDGVPVVTSAKITEL
MMAR_4289|M.marinum_M LVTNLDASAFLGRLALVRIYNGKLRKGQQVAWLREVDGVPVVTSAKITEL
MLBr_01498|M.leprae_Br4923 LVTNLDASAFLGRLALVRIYNGKLRKGQQVAWMREVDGLPVTTDAKITEL
Mflv_2163|M.gilvum_PYR-GCK LVTNLDASAFLGRLALIRIYKGRIKKGQQVAWMREVDGHPVITNAKITEL
Mvan_4540|M.vanbaalenii_PYR-1 LVTNLDASAFLGRLALIRIYKGRIKKGQQVAWMREVDGHPVITTAKITEL
MSMEG_5132|M.smegmatis_MC2_155 LVTNLDASAFLGRLALVRIYNGKLKKGQQVAWMREVDGAPVVTSAKITEL
MAB_1310|M.abscessus_ATCC_1997 LVTNLDASAFLGRLALIRIYNGRIRKGQQVAWMREVDGHPVITNAKITEL
MAV_1307|M.avium_104 LVTNLDASTFLGRLALIRIYNGKLCKGQQVAWMREVDGLPVITSAKITEL
TH_1877|M.thermoresistible__bu LVTNLDASSFLGRLALVRIANGRLRKGQQVAWIREGE----TTQAKITEL
********:*******:** :*:: *******:*: : * ******
Mb1197|M.bovis_AF2122/97 LATEGVERKPTDAAVAGDIVAVAGLPEIMIGDTLAASANPVALPRITVDE
Rv1165|M.tuberculosis_H37Rv LATEGVERKPTDAAVAGDIVAVAGLPEIMIGDTLAASANPVALPRITVDE
MUL_1009|M.ulcerans_Agy99 LATEGVERSTTDEAVAGDIVAVAGLPEIMIGDTLADPDHAHALPRITVDE
MMAR_4289|M.marinum_M LATEGVERSTTDEAVAGDIVAVAGLPEIMIGDTLADPDHAHALPRITVDE
MLBr_01498|M.leprae_Br4923 LVTKGVERSTTEEATAGDIVAVAGLPEIMIGDTLADPEHAHALPRITVDE
Mflv_2163|M.gilvum_PYR-GCK LVTVGVERTATDEAFAGDIVAVAGIPEIMIGDTLADPDHAHALPRITVDE
Mvan_4540|M.vanbaalenii_PYR-1 LVTEGVERTATDEAFAGDIVAVAGIPEIMIGDTLADPDHAHALPRITVDE
MSMEG_5132|M.smegmatis_MC2_155 LVTEGVERTATAEAVAGDIVAVAGIPEIMIGDTLADPDHAHALPRITVDE
MAB_1310|M.abscessus_ATCC_1997 LATEGVDRSPTEEAVAGDIVAVAGMSEIMIGDTLADPEHAHALPRITVDE
MAV_1307|M.avium_104 LATEGVERSPTDEAIAGDIVAVAGLPEIMIGDTLADPDHAHALPRITVDE
TH_1877|M.thermoresistible__bu LVTVGVERSPTTEAQAGDIVAVAGLPDIMIGDTLADPEHPVALPRIAVDE
*.* **:*..* * *********:.:******** . :. *****:***
Mb1197|M.bovis_AF2122/97 PAISVTIGTNTSPLAGKVGGHKLTARMVRSRLDAELVGNVSIRVVDIGAP
Rv1165|M.tuberculosis_H37Rv PAISVTIGTNTSPLAGKVGGHKLTARMVRSRLDAELVGNVSIRVVDIGAP
MUL_1009|M.ulcerans_Agy99 PAISVTVGTNTSPLAGKVSGHKLTARMVRGRLDTELIGNVSIRVVDIGRP
MMAR_4289|M.marinum_M PAISVTVGTNTSPLAGKVSGHKLTARMVRGRLDTELIGNVSIRVVDIGRP
MLBr_01498|M.leprae_Br4923 PAISVTIGTNTSPLAGKVSGHKLTARMVRGRLDAELVGNISIRVVDIGRP
Mflv_2163|M.gilvum_PYR-GCK PAISVTIGTNTSPLAGKVSGHKLTARMVKNRLDSELVGNVSIKVVDIGRP
Mvan_4540|M.vanbaalenii_PYR-1 PAISVTIGTNTSPLAGKVSGHKLTARMVKNRLDSELVGNVSIKVVDIGRP
MSMEG_5132|M.smegmatis_MC2_155 PAISVTIGTNTSPLAGKVSGHKLTARMVKNRLDSELIGNVSIRVVDIGKP
MAB_1310|M.abscessus_ATCC_1997 PAISVTIGTNSSPLAGKVSGHKLTARMVKSRLDSELIGNVSVKVVDIGRP
MAV_1307|M.avium_104 PAISVTIGTNTSPLAGKVPGHKLTARLVRNRLDQELVGNVSIRVVDIGRP
TH_1877|M.thermoresistible__bu PAISVTIGTNTSPLAGKVSGHKLTARMVKSRLDSELVGNVSIRVVDIGRP
******:***:******* *******:*:.*** **:**:*::***** *
Mb1197|M.bovis_AF2122/97 DAWEVQGRGELALAVLVEQMRREGFELTVGKPQVVTKTIDGTLHEPFESM
Rv1165|M.tuberculosis_H37Rv DAWEVQGRGELALAVLVEQMRREGFELTVGKPQVVTKTIDGTLHEPFESM
MUL_1009|M.ulcerans_Agy99 DAWEVQGRGELALAVLVETMRREGFELTVGKPQVVTKTIDGQLHEPFEAM
MMAR_4289|M.marinum_M DAWEVQGRGELALAVLVETMRREGFELTVGKPQVVTKTIDGQLHEPFEAM
MLBr_01498|M.leprae_Br4923 DAWEVQGRGELALAVLVETMRREGFELTVGKPQVVTRTIDGKLHEPFEVM
Mflv_2163|M.gilvum_PYR-GCK DAWEVQGRGELALAVLVEQMRREGFELTVGKPQVVTRTIDGKLHEPFEAM
Mvan_4540|M.vanbaalenii_PYR-1 DAWEVQGRGELALAVLVEQMRREGFELTVGKPQVVTRTVDGKLHEPFEAM
MSMEG_5132|M.smegmatis_MC2_155 DAWEVQGRGELALAVLVEQMRREGFELTVGKPQVVTKNIDGKLHEPFEAM
MAB_1310|M.abscessus_ATCC_1997 DAWEVQGRGELALAILVEQMRREGFELTVGKPQVVTRQIDGKLHEPFEAM
MAV_1307|M.avium_104 DAWEVQGRGELALAVLVEQMRREGFELTVGKPQVVTRTIDGQLHEPFEAM
TH_1877|M.thermoresistible__bu DAWEVQGRGELALAVLVETMRREGFELTVGKPQVVTKTIDGKLHEPYEAL
**************:*** *****************: :** ****:* :
Mb1197|M.bovis_AF2122/97 TVDCPEEYIGAVTQLMAARKGRMVEMANHTTGWVRMDFVVPSRGLIGWRT
Rv1165|M.tuberculosis_H37Rv TVDCPEEYIGAVTQLMAARKGRMVEMANHTTGWVRMDFVVPSRGLIGWRT
MUL_1009|M.ulcerans_Agy99 TIDCPDEFVGAITQLMAGRKGRVEEMTNHAAGWVRMDFIVPSRGLIGFRT
MMAR_4289|M.marinum_M TIDCPDEFVGAITQLMAGRKGRMEEMTNHAAGWVRMDFIVPSRGLIGFRT
MLBr_01498|M.leprae_Br4923 TIDCPEEFVGAITQLMAGRKGRMEEMANHAVGWVRMDFIVPSRGLIGFRT
Mflv_2163|M.gilvum_PYR-GCK TIDCPEEFVGAITQLMAARKGRMEEMTNHAAGWVRMDFIVPSRGLIGFRT
Mvan_4540|M.vanbaalenii_PYR-1 TIDCPEEFVGAITQLMAARKGRMEEMTNHAAGWVRMDFIVPSRGLIGFRT
MSMEG_5132|M.smegmatis_MC2_155 TIDCPEEFVGAITQLMAARKGRMEEMTNHAAGWVRMDFIVPSRGLIGFRT
MAB_1310|M.abscessus_ATCC_1997 TIDCPEEFVGAITQLMAARKGRMEEMTNHAAGWVRMDFIVPSRGLIGFRT
MAV_1307|M.avium_104 TIDCPEEFVGAITQLMAARKGRMEEMTNHAAGWVRMDFIVPSRGLIGFRT
TH_1877|M.thermoresistible__bu TIDCPEEFVGSITQMMAARKGRMEHMVNHAAGWVRMDYTIPSRGLIGFRT
*:***:*::*::**:**.****: .*.**:.******: :*******:**
Mb1197|M.bovis_AF2122/97 DFLTETRGSGVGHAVFDGYRPWAGEIRARHTGSLVSDRAGAITPFALLQL
Rv1165|M.tuberculosis_H37Rv DFLTETRGSGVGHAVFDGYRPWAGEIRARHTGSLVSDRAGAITPFALLQL
MUL_1009|M.ulcerans_Agy99 DFLTLTRGTGIANAVFDGYRPWAGEIRARHTGSLVSDRSGVITPFALLQL
MMAR_4289|M.marinum_M DFLTLTRGTGIANAVFDGYRPWAGEIRARHTGSLVSDRSGAITPFALLQL
MLBr_01498|M.leprae_Br4923 DFLTLTRGTGIANAVFDSYRPWAGEIRARDTGSLVSDRPGTITPFALLQL
Mflv_2163|M.gilvum_PYR-GCK DFLTLTRGTGIANAVFDGYRPWAGEIRARHTGSLVSDRSGQITPFAMIQL
Mvan_4540|M.vanbaalenii_PYR-1 DFLTLTRGTGIANAVFDGYRPWAGEIRARHTGSLVSDRSGQITPFAMIQL
MSMEG_5132|M.smegmatis_MC2_155 DFLTLTRGTGIANAVFDGYRPWAGEIRARHTGSLVSDRAGTITPFAMIQL
MAB_1310|M.abscessus_ATCC_1997 DFLTLTRGTGIANAVFEGYRPWAGEIRARHTGSLVSDRSGTITPFAMIQL
MAV_1307|M.avium_104 DFLTITRGTGIANAVFDGYRPWAGEIRARHTGSLVSDRAGTITPFAMIQL
TH_1877|M.thermoresistible__bu DFLTETRGTGIAHAVFDGYRPWAGEIRARRSGSLVSDRAGQVTAYALLQL
**** ***:*:.:***:.*********** :*******.* :*.:*::**
Mb1197|M.bovis_AF2122/97 ADRGQFFVEPGQQTYEGMVVGINPRPEDLDINVTREKKLTNMRSSTADVI
Rv1165|M.tuberculosis_H37Rv ADRGQFFVEPGQQTYEGMVVGINPRPEDLDINVTREKKLTNMRSSTADVI
MUL_1009|M.ulcerans_Agy99 ADRGQFFVEPGEDTYEGMVVGINPRAEDLDINVTREKKLTNMRSSTADVI
MMAR_4289|M.marinum_M ADRGQFFVEPGEDTYEGMVVGINPRAEDLDINVTREKKLTNMRSSTADVI
MLBr_01498|M.leprae_Br4923 ADRGQFFVSPGQDTYQGMVVGINPRPEDLDINVTREKKLTNMRSSTANVI
Mflv_2163|M.gilvum_PYR-GCK ADRGQFFVEPGQETYEGQVVGINPRAEDLDINITREKKLTNMRSSTADVM
Mvan_4540|M.vanbaalenii_PYR-1 ADRGQFFVEPGQETYEGQVVGINPRAEDLDINVTREKKLTNMRSSTADVM
MSMEG_5132|M.smegmatis_MC2_155 ADRGQFFVEPGQETYEGMVVGINPRAEDLDINVTREKKLTNMRSSTADVM
MAB_1310|M.abscessus_ATCC_1997 ADRGQFFVEPGEDTYEGMVVGINPRAEDLDVNVTREKKLTNMRSSTADVM
MAV_1307|M.avium_104 ADRGQFFVEPGQDTYEGMVVGINPRAEDLDINVTREKKLTNMRSSTADVI
TH_1877|M.thermoresistible__bu ADRGTFFVEPGDETYEGMVVGVNPRAEDLDVNVTREKKLTNMRASTAEVM
**** ***.**::**:* ***:***.****:*:**********:***:*:
Mb1197|M.bovis_AF2122/97 ETLAKPLQLDLERAMELCAPDECVEVTPEIVRIRKVELAAAARARSRART
Rv1165|M.tuberculosis_H37Rv ETLAKPLQLDLERAMELCAPDECVEVTPEIVRIRKVELAAAARARSRART
MUL_1009|M.ulcerans_Agy99 ETLAKPLELDLERAIEFCAPDECVEVTPEAIRVRKVELDATSRARSRSRA
MMAR_4289|M.marinum_M ETLAKPLELDLERAMEFCAPDECVEVTPEAIRVRKVELDATSRARSRSRA
MLBr_01498|M.leprae_Br4923 ETLTKPLELDLERAMEFCSFDECVEVTPEIVRVRKIELESNARARGRARA
Mflv_2163|M.gilvum_PYR-GCK ETLARPLELDLEKAMEFCAEDECVEVTPEIVRVRKVELTASLRARAKARA
Mvan_4540|M.vanbaalenii_PYR-1 ETLARPLELDLEKAMEFCAADECVEVTPEIVRVRKVELTASLRARAKSRA
MSMEG_5132|M.smegmatis_MC2_155 ETLARPLELDLEKAMEFCAEDECVEVTPEVVRVRKVELTASLRARAKARA
MAB_1310|M.abscessus_ATCC_1997 ETLARPIELDLEQAMEFCAADECVEVTPEIVRVRKVDLDANTRARNRSRA
MAV_1307|M.avium_104 ETLAKPLELDLEQAMEFCAADECVEVTPEIVRVRKVELDATSRARSRARA
TH_1877|M.thermoresistible__bu ETLAKPIQLDLERAMEFCAADECVEVTPESIRVRKVELSATARARARSRA
***::*::****:*:*:*: ********* :*:**::* : *** ::*:
Mb1197|M.bovis_AF2122/97 KARG---
Rv1165|M.tuberculosis_H37Rv KARG---
MUL_1009|M.ulcerans_Agy99 KARG---
MMAR_4289|M.marinum_M KARG---
MLBr_01498|M.leprae_Br4923 KVRG---
Mflv_2163|M.gilvum_PYR-GCK KARG---
Mvan_4540|M.vanbaalenii_PYR-1 KQANN--
MSMEG_5132|M.smegmatis_MC2_155 KARG---
MAB_1310|M.abscessus_ATCC_1997 KAAANNS
MAV_1307|M.avium_104 KARG---
TH_1877|M.thermoresistible__bu KARA---
*