For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRLFAHTVRPAMLAASRADRLRRTLEHSSVTRRVVRRFVPGETVDSVLDVVGALRDSGRCVNIDYLGEN VTDVDDAAQTVRVYLNLLDVMGRRNDAGRDGVRPLEVSVKLSALGQALERDGQKIALDNARTICERAEQV GVWVTVDAEDHTTTESTLSIAGDLRVDFAWLGTVVQAYLRRTLDDCQELASVGARVRLCKGAYDEPESVA YRDRVEVTDSYLRCLQVLMAGSGYPMVASHDPEIIATVAGMAHESGRSANDFEYQMLYGIRDEEQWRLVG AGSQVRVYVPFGTQWYGYFMRRLAERPANLGFFLRALVK
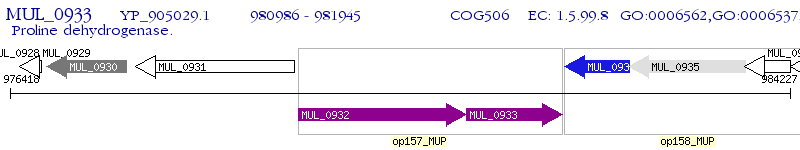
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0933 | putA | - | 100% (319) | proline dehydrogenase, PutA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1220 | - | 1e-143 | 78.55% (317) | proline dehydrogenase |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv1188 | - | 1e-143 | 78.55% (317) | proline dehydrogenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1331 | - | 1e-115 | 63.21% (318) | proline dehydrogenase |
| M. marinum M | MMAR_4252 | putA | 0.0 | 99.06% (319) | proline dehydrogenase, PutA |
| M. avium 104 | MAV_1332 | - | 1e-123 | 69.72% (317) | proline dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5117 | - | 1e-116 | 67.41% (316) | proline dehydrogenase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4525 | - | 1e-120 | 67.72% (316) | proline dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5117|M.smegmatis_MC2_155 ---MGIFDRVARPAILAASRSQRLRATAERLPVTRRVVDRFVAGETVPEA
Mvan_4525|M.vanbaalenii_PYR-1 ---MSVFDKIARPVILAASRREGLRRAAERMSVTRKVVRRFVPGESTADV
MAB_1331|M.abscessus_ATCC_1997 MRMAEWFGKTARPAILAASRSARLRHAAERMPLTRQVVRRFVPGESQDDV
MUL_0933|M.ulcerans_Agy99 --MTRLFAHTVRPAMLAASRADRLRRTLEHSSVTRRVVRRFVPGETVDSV
MMAR_4252|M.marinum_M --MTRLFAHTVRPAMLAASRADRLRRTLEHSSVTRRVVRRFVPGETVDSV
Mb1220|M.bovis_AF2122/97 --MAGWFAHTLRPAMLAAGRSDRLGRIVERSPLTRGVVRRFVPGDTLDDV
Rv1188|M.tuberculosis_H37Rv --MAGWFAHTLRPAMLAAGRSDRLGRIVERSPLTRGVVRRFVPGDTLDDV
MAV_1332|M.avium_104 --MAGLFAHTLRPAILAAGRRPGLRRAAEALPVTRRVVHRFIAGETIDSA
* : **.:***.* * * .:** ** **:.*:: ..
MSMEG_5117|M.smegmatis_MC2_155 LDATAALRASGRLVTIDYLGEDTTSKEDADNTVAAYLALLEGLKS--DSD
Mvan_4525|M.vanbaalenii_PYR-1 ITAVGDLRDSHRLVSIDYLGEDVTDADAADATVEAYLTLLDALAGRGETA
MAB_1331|M.abscessus_ATCC_1997 LGASTGLLDSGRFISIDYLGEDTTDSEQATRTVSAYLSLLDALAQRSDAV
MUL_0933|M.ulcerans_Agy99 LDVVGALRDSGRCVNIDYLGENVTDVDDAAQTVRVYLNLLDVMGRRNDAG
MMAR_4252|M.marinum_M LDVVGALRDSGRCVSIDYLGENVTDVDDAAETVRAYLNLLDVMGRRNDAG
Mb1220|M.bovis_AF2122/97 VDIVTALRDSGRYLSIDYLGENVTDADDAAAAVRAYLGLLDVLGRRGDIA
Rv1188|M.tuberculosis_H37Rv VDIVTALRDSGRYLSIDYLGENVTDADDAAAAVRAYLGLLDVLGRRGDIA
MAV_1332|M.avium_104 LDSVTALRDSGRCVSVDHLGEDVSDADDADAAVRVYLELVDRLGCLGDG-
: * * * :.:*:***:.:. : * :* .** *:: : :
MSMEG_5117|M.smegmatis_MC2_155 AK-VRPLEVSLKLSALGQALPRDGEKIALENAHTICAKARDIEAWVTVDA
Mvan_4525|M.vanbaalenii_PYR-1 AA-VRPLEVSLKLSALGQALPRDGEKIALENARAICERAQRAGVWVTVDA
MAB_1331|M.abscessus_ATCC_1997 HTGVRPLEISLKLSALGQALPRDGERIALENAQLLCAKADQAGVWVTADA
MUL_0933|M.ulcerans_Agy99 RDGVRPLEVSVKLSALGQALERDGQKIALDNARTICERAEQVGVWVTVDA
MMAR_4252|M.marinum_M RDGVRPLEVSVKLSALGQALERDGQKIALDNARTICERAEQVGVWVTVDA
Mb1220|M.bovis_AF2122/97 CDGVRPLEVSLKLSALGQALDRDGQKIALDNARAICERAERVGAWVTVDA
Rv1188|M.tuberculosis_H37Rv CDGVRPLEVSLKLSALGQALDRDGQKIALDNARAICERAERVGAWVTVDA
MAV_1332|M.avium_104 -AAVRPLEVSLKLSALGQSLDRDGEKIARENAWAICAAAQRAGVWVTVDA
*****:*:*******:* ***::** :** :* * .***.**
MSMEG_5117|M.smegmatis_MC2_155 EDHTTTDSTLSIVRDLRTEFDWLGTVLQAYLKRTRADCAEFAASGARIRL
Mvan_4525|M.vanbaalenii_PYR-1 EDHTTTDSTLAIVRELRVDFDWVGTVLQSHLKRARADCAEFAASGARIRL
MAB_1331|M.abscessus_ATCC_1997 EDHTTTDSTLNIVRELRADFPGLGTVLQAYLKRTYADCVDLSGKGSRIRL
MUL_0933|M.ulcerans_Agy99 EDHTTTESTLSIAGDLRVDFAWLGTVVQAYLRRTLDDCQELASVGARVRL
MMAR_4252|M.marinum_M EDHTTTESTLSIAGDLRVDFAWLGTVVQAYLRRTLDDCQELASVGARVRL
Mb1220|M.bovis_AF2122/97 EDHTTTDSTLSISGDLRVDFPWLGTVVQAYLRRTLADCAELAAVGARVRL
Rv1188|M.tuberculosis_H37Rv EDHTTTDSTLSISGDLRVDFPWLGTVVQAYLRRTLADCAELAAVGARVRL
MAV_1332|M.avium_104 EGHTATDSTLRIVRDLRREFDWLGVALQAYLRRTLGDCEEFAAAGARVRL
*.**:*:*** * :** :* :*..:*::*:*: ** :::. *:*:**
MSMEG_5117|M.smegmatis_MC2_155 CKGAYDEPASVAYRDPDEVTDSYLTCLRILMAGRGYPMVASHDPEIIAAV
Mvan_4525|M.vanbaalenii_PYR-1 CKGAYAEPASVAFRDRDDVTDSYLQCLRILMAGRGYPMVASHDPEILGAV
MAB_1331|M.abscessus_ATCC_1997 CKGAYDEPAAVAHRDPEDVDVAYLRCLRVLMEGDGYPMVASHDPAIIEAA
MUL_0933|M.ulcerans_Agy99 CKGAYDEPESVAYRDRVEVTDSYLRCLQVLMAGSGYPMVASHDPEIIATV
MMAR_4252|M.marinum_M CKGAYDEPESVAYRDRVEVTDSYLRCLQVLMAGSGYPMVASHDPEIIATV
Mb1220|M.bovis_AF2122/97 CKGAYDEPASVAYRDAAQVTDSYLRCLRVLTAGRGYPMVATHDPVIIAAV
Rv1188|M.tuberculosis_H37Rv CKGAYDEPASVAYRDAAQVTDSYLRCLRVLTAGRGYPMVATHDPVIIAAV
MAV_1332|M.avium_104 CKGAYDEPASLAYRDPAAVTDSYLRCLRVLMAGSGYPMVASHDPAIIEAA
***** ** ::*.** * :** **::* * ******:*** *: :.
MSMEG_5117|M.smegmatis_MC2_155 PALAREYHRGVDDFEYQMLYNIRDGEQRRLADEGNHVRVYVPFGYEWYGY
Mvan_4525|M.vanbaalenii_PYR-1 PSMAREYGRGVDDFEYQMLYGIRDAEQRRLAEAGNHIRVYVPFGTQWYGY
MAB_1331|M.abscessus_ATCC_1997 GLMARETGRGPGDYEYQMLYGIREAEQRRLAGVGHHMRVYVPFGEQWYGY
MUL_0933|M.ulcerans_Agy99 AGMAHESGRSANDFEYQMLYGIRDEEQWRLVGAGSQVRVYVPFGTQWYGY
MMAR_4252|M.marinum_M AGMAHESGRSANDFEYQMLYGIRDEEQWRLVGAGSQVRVYVPFGTQWYGY
Mb1220|M.bovis_AF2122/97 PGITRESGRSQGDFEYQMLYGVRDDEQRRLTGAGNHVRVYVPFGTRWYGY
Rv1188|M.tuberculosis_H37Rv PGITRESGRSQGDFEYQMLYGVRDDEQRRLTGAGNHVRVYVPFGTRWYGY
MAV_1332|M.avium_104 PSLARESGRGTGEFEYQMLYGIRDDEQRRLAEAGHTVRVYVPFGTQWYGY
:::* *. .::******.:*: ** **. * :******* .****
MSMEG_5117|M.smegmatis_MC2_155 FVRRLAERPANLTFFLRALAERR--------
Mvan_4525|M.vanbaalenii_PYR-1 FVRRLAERPANLTFFLRALAERRG-------
MAB_1331|M.abscessus_ATCC_1997 FVRRIAERPANMTFFLRALVSGG--------
MUL_0933|M.ulcerans_Agy99 FMRRLAERPANLGFFLRALVK----------
MMAR_4252|M.marinum_M FMRRLAERPANLGFFLRALVK----------
Mb1220|M.bovis_AF2122/97 FLRRLAERPANLAFFLRALTDRRRARGCAER
Rv1188|M.tuberculosis_H37Rv FLRRLAERPANLAFFLRALTDRRRARGCAER
MAV_1332|M.avium_104 FMRRLAERPANLTFFLRALAARRH-------
*:**:******: ******.