For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTTTEFPSATAKARFVRVSPRKARRVIDLVRGRSVSDALDILRWAPQAASEPVAKVIASAAANAQNNNG LDPATLIVATVYADEGPTAKRIRPRAQGRAFRIRKRTSHITVVVESRPSKDQRSSKSSRARRAEGSKAAA TAPAKKSSASKAPAKKAATKAESKTSEISEAKGGSD
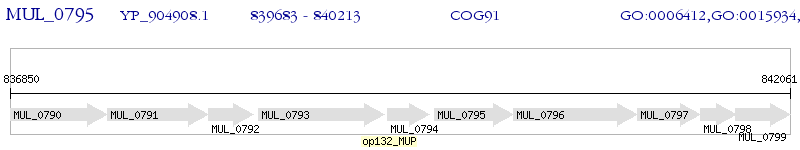
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0795 | rplV | - | 100% (176) | 50S ribosomal protein L22 |
| M. ulcerans Agy99 | MUL_1970 | hupB | 4e-05 | 51.22% (41) | DNA-binding protein HU HupB-like protein |
| M. ulcerans Agy99 | MUL_3764 | plsC | 7e-05 | 38.33% (60) | bifunctional transmembrane phospholipid biosynthesis enzyme |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0726 | rplV | 5e-75 | 78.87% (194) | 50S ribosomal protein L22 |
| M. gilvum PYR-GCK | Mflv_5044 | rplV | 3e-61 | 83.69% (141) | ribosomal protein L22 |
| M. tuberculosis H37Rv | Rv0706 | rplV | 5e-75 | 78.87% (194) | 50S ribosomal protein L22 |
| M. leprae Br4923 | MLBr_01858 | rplV | 5e-76 | 82.76% (174) | 50S ribosomal protein L22 |
| M. abscessus ATCC 19977 | MAB_3815c | rplV | 2e-62 | 79.87% (154) | 50S ribosomal protein L22 |
| M. marinum M | MMAR_1036 | rplV | 4e-92 | 99.43% (176) | 50S ribosomal protein L22, RplV |
| M. avium 104 | MAV_4466 | rplV | 1e-81 | 87.71% (179) | 50S ribosomal protein L22 |
| M. smegmatis MC2 155 | MSMEG_1441 | rplV | 8e-63 | 81.58% (152) | 50S ribosomal protein L22 |
| M. thermoresistible (build 8) | TH_0402 | rplV | 4e-61 | 73.26% (172) | PROBABLE 50S RIBOSOMAL PROTEIN L22 RPLV |
| M. vanbaalenii PYR-1 | Mvan_1310 | rplV | 2e-57 | 73.55% (155) | ribosomal protein L22 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1441|M.smegmatis_MC2_155 --MST-VTEFPSATAKARYVRVSATKARRVIDLVRGKSVEEALDILRWAP
TH_0402|M.thermoresistible__bu --MSAPVIEYPSATAKARFVRTSATKARRVIDLVRGKTVAEALDILRWAP
Mflv_5044|M.gilvum_PYR-GCK --MTT-AIQYPSASAKARFVRVSPTKARRVIDLVRGKSVEDALDILRWAP
Mvan_1310|M.vanbaalenii_PYR-1 --MTT-AIEYPSAMAKARFVRISATKARRVIDLVRGKSVEEALDILRWAP
MAB_3815c|M.abscessus_ATCC_199 --MTT-TTEFPSATAKARFVRVSPTKARRVIDLVRGKSVNDAIDILRWAP
MUL_0795|M.ulcerans_Agy99 ---MTTTTEFPSATAKARFVRVSPRKARRVIDLVRGRSVSDALDILRWAP
MMAR_1036|M.marinum_M ---MTTTTEFPSATAKARFVRVSPRKARRVIDLVRGRSVSDALDILRWAP
MAV_4466|M.avium_104 ----MTTTEFPSAVAKARFVRVSPRKARRVIDLVRGRSVTDALDILRWAP
MLBr_01858|M.leprae_Br4923 ----MTTTEFPSAIAKARFVRVSPRKARRVIDLVRGRSVADALDILRWAP
Mb0726|M.bovis_AF2122/97 MTAATKATEYPSAVAKARFVRVSPRKARRVIDLVRGRSVSDALDILRWAP
Rv0706|M.tuberculosis_H37Rv MTAATKATEYPSAVAKARFVRVSPRKARRVIDLVRGRSVSDALDILRWAP
. ::*** ****:** *. ***********::* :*:*******
MSMEG_1441|M.smegmatis_MC2_155 QAASEPVAKVIASAAANAQNNEGLDPSTLVVATVYADEGPTAKRIRPRAQ
TH_0402|M.thermoresistible__bu QAASEPVAKVIASAAANAQNNEGLDPDTLVVATIYADEGPTFKRIRPRAQ
Mflv_5044|M.gilvum_PYR-GCK QAASEPVAKVIASAAANAQNNEGLDPSTLVVATVFADEGPTAKRIRPRAQ
Mvan_1310|M.vanbaalenii_PYR-1 QSASEPVAKVIASAAANAQNNEGLDPTSLVVATIHADEGPTAKRIRPRAQ
MAB_3815c|M.abscessus_ATCC_199 QAASEPVAKVIASAAANAQNNDGLDPSTLVVSEIFADEGPTAKRIRPRAQ
MUL_0795|M.ulcerans_Agy99 QAASEPVAKVIASAAANAQNNNGLDPATLIVATVYADEGPTAKRIRPRAQ
MMAR_1036|M.marinum_M QAASEPVAKVIASAAANAQNNNGLDPATLIVATVYADEGPTAKRIRPRAQ
MAV_4466|M.avium_104 QAASEPVAKVIASAAANAQNNNGLDPATLVVATVYADEGPTAKRIRPRAQ
MLBr_01858|M.leprae_Br4923 QAASEPVARVIASAAANAQNNNGLDPETLVVATVYAGEGPTAKRIRPRAQ
Mb0726|M.bovis_AF2122/97 QAASGPVAKVIASAAANAQNNGGLDPATLVVATVYADQGPTAKRIRPRAQ
Rv0706|M.tuberculosis_H37Rv QAASGPVAKVIASAAANAQNNGGLDPATLVVATVYADQGPTAKRIRPRAQ
*:** ***:************ **** :*:*: :.*.:*** ********
MSMEG_1441|M.smegmatis_MC2_155 GRAFRIRKRTSHITVIVESRPPKQ----KGASAASARSRRAQGSKA----
TH_0402|M.thermoresistible__bu GRAFRIRRRTSHITVVVESRPVKKG---RGGSAASARSRRAQASKA----
Mflv_5044|M.gilvum_PYR-GCK GRAFRIRKRTSHITVIVESRPAK----KQGASASAARARRAQASK-----
Mvan_1310|M.vanbaalenii_PYR-1 GRAYRIRKRTSHITVIVESRPPKKAG-KQGASASAARARRAQASK-----
MAB_3815c|M.abscessus_ATCC_199 GRAFRIRKRTSHITVVVESRPKQEKGGKAGASKASSRAARAQGSKA----
MUL_0795|M.ulcerans_Agy99 GRAFRIRKRTSHITVVVESRPSKDQ-----RSSKSSRARRAEGS------
MMAR_1036|M.marinum_M GRAFRIRKRTSHITVVVESRPSKDQ-----RSSKSSRARRAEGS------
MAV_4466|M.avium_104 GRAFRIRKRTSHITVVVESRPAKDQ-----RSAKSSRTRRAEAS------
MLBr_01858|M.leprae_Br4923 GRAFRIRRRTSHITVVVESRPVKDQ-----RSAASTKARRAEAS------
Mb0726|M.bovis_AF2122/97 GRAFRIRRRTSHITVVVESRPAKDQ-----RSAKSSRARRTEASKAASKV
Rv0706|M.tuberculosis_H37Rv GRAFRIRRRTSHITVVVESRPAKDQ-----RSAKSSRARRTEASKAASKV
***:***:*******:***** : * :::: *::.*
MSMEG_1441|M.smegmatis_MC2_155 ------------AATKKSAETKE--------------------------G
TH_0402|M.thermoresistible__bu ------------ASTKKAPATKATAQKA--------------TADEAKGG
Mflv_5044|M.gilvum_PYR-GCK ------------AASQ-----KE--------------------------G
Mvan_1310|M.vanbaalenii_PYR-1 ------------AATKKATDSKE--------------------------G
MAB_3815c|M.abscessus_ATCC_199 ------------AAAKKT-ESKG--------------------------G
MUL_0795|M.ulcerans_Agy99 -------KAAATAPAKKSSASKAPAKKA-----ATKAESKTSEISEAKGG
MMAR_1036|M.marinum_M -------KAAATAPAKKSSASKAPAKKA-----ATKAESKTSETSEAKGG
MAV_4466|M.avium_104 -------KAAAKAPAKKAPAKKAPAKKAPAKTAAKKTPAKTSETSDAKGG
MLBr_01858|M.leprae_Br4923 -------KTAGRAPAKKAGASSGATKMP-----PKKASVKTSEASETKGG
Mb0726|M.bovis_AF2122/97 GATAPAKKAAAKAPAKKAPASSGVKKTPAKKAPAKKAPAKASETSAAKGG
Rv0706|M.tuberculosis_H37Rv GATAPAKKAAAKAPAKKAPASSGVKKTPAKKAPAKKAPAKASETSAAKGG
*.:: . *
MSMEG_1441|M.smegmatis_MC2_155 SE
TH_0402|M.thermoresistible__bu SQ
Mflv_5044|M.gilvum_PYR-GCK SE
Mvan_1310|M.vanbaalenii_PYR-1 SE
MAB_3815c|M.abscessus_ATCC_199 TS
MUL_0795|M.ulcerans_Agy99 SD
MMAR_1036|M.marinum_M SD
MAV_4466|M.avium_104 SD
MLBr_01858|M.leprae_Br4923 SD
Mb0726|M.bovis_AF2122/97 SD
Rv0706|M.tuberculosis_H37Rv SD
:.