For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRGARVADTGGERRPVAFIAMTDPEKSPESSGRGPASQGVTYAAAGVDIEAGDRAVELFKPLATRATRPE VRGGLGGFAGLFALRNDYREPVLAASTDGVGTKLAVAQAMDKHDTVGLDLVAMVVDDLVVCGAEPLFLQD YIAVGRTVPERVSAIVSGIAEGCVRAGCALLGGETAEHPGLMAPDHYDISATGVGVVEADNVLGPERVKP GDVIIAMGSSGLHSNGYSLARNVLLEIDRMNLAGYVEEFGRTLGEELLEPTRIYAKDCLALTAETHVRTF CHVTGGGLAGNLERVIPHGLVAEIDRGTWTPAPVFAMIAQRGRVVREEMEKTFNMGVGMIAVVAPEDTDR ALAILTARHLDCWVLGTVCKGGKEGPRATLVGQHPRF
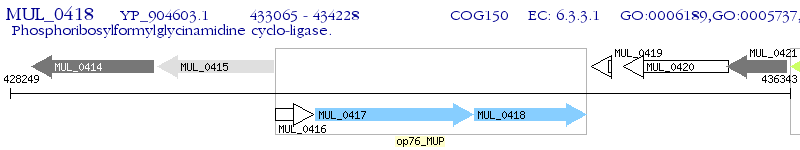
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0418 | purM | - | 100% (387) | phosphoribosylaminoimidazole synthetase |
| M. ulcerans Agy99 | MUL_1978 | thiL | 8e-06 | 27.13% (129) | thiamine monophosphate kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0832 | purM | 0.0 | 88.01% (367) | phosphoribosylaminoimidazole synthetase |
| M. gilvum PYR-GCK | Mflv_1636 | - | 0.0 | 85.91% (362) | phosphoribosylaminoimidazole synthetase |
| M. tuberculosis H37Rv | Rv0809 | purM | 0.0 | 88.01% (367) | phosphoribosylaminoimidazole synthetase |
| M. leprae Br4923 | MLBr_02205 | purM | 0.0 | 88.28% (367) | phosphoribosylaminoimidazole synthetase |
| M. abscessus ATCC 19977 | MAB_0728 | - | 1e-168 | 82.81% (349) | phosphoribosylaminoimidazole synthetase |
| M. marinum M | MMAR_4880 | purM | 0.0 | 99.73% (367) | 5'-phosphoribosyl-5-aminoimidazole synthetase, PurM |
| M. avium 104 | MAV_0748 | purM | 0.0 | 92.92% (367) | phosphoribosylaminoimidazole synthetase |
| M. smegmatis MC2 155 | MSMEG_5798 | purM | 0.0 | 86.92% (367) | phosphoribosylaminoimidazole synthetase |
| M. thermoresistible (build 8) | TH_0070 | purM | 0.0 | 87.97% (349) | PROBABLE PHOSPHORIBOSYLFORMYLGLYCINAMIDINE CYCLO-LIGASE PURM |
| M. vanbaalenii PYR-1 | Mvan_5111 | - | 0.0 | 83.86% (378) | phosphoribosylaminoimidazole synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0418|M.ulcerans_Agy99 MRGARVADTGGERRPVAFIAMTDPEKSPESS----------------GRG
MMAR_4880|M.marinum_M --------------------MTDPEKSPESS----------------GRG
MAV_0748|M.avium_104 --------------------MTDPGKSP-------------------GRH
Mb0832|M.bovis_AF2122/97 --------------------MTDLAKGP-------------------GKD
Rv0809|M.tuberculosis_H37Rv --------------------MTDLAKGP-------------------GKD
MLBr_02205|M.leprae_Br4923 --------------------MMDPGKSP-------------------GGQ
MSMEG_5798|M.smegmatis_MC2_155 --------------------MTDREDSV----------------------
Mflv_1636|M.gilvum_PYR-GCK --------------------MLSMTKGVEQ-----------------SGI
Mvan_5111|M.vanbaalenii_PYR-1 -------------------MAGSPTTCAERTPGRIGCSRRPVALWPMSDR
TH_0070|M.thermoresistible__bu --------------------MTER--------------------------
MAB_0728|M.abscessus_ATCC_1997 --------------------MPDMSER-----------------------
.
MUL_0418|M.ulcerans_Agy99 PASQGVTYAAAGVDIEAGDRAVELFKPLATRATRPEVRGGLGGFAGLFAL
MMAR_4880|M.marinum_M PASQGVTYAAAGVDIEAGDRAVELFKPLATRATRPEVRGGLGGFAGLFAL
MAV_0748|M.avium_104 SGSQGITYASAGVDIEAGDRAVELFKPLATKATRPEVRGGLGGFAGLFAL
Mb0832|M.bovis_AF2122/97 PGSRGITYASAGVDIEAGDRAIDLFKPLASKATRPEVRGGLGGFAGLFTL
Rv0809|M.tuberculosis_H37Rv PGSRGITYASAGVDIEAGDRAIDLFKPLASKATRPEVRGGLGGFAGLFTL
MLBr_02205|M.leprae_Br4923 LGNHGITYASTGVDIEAGDRSVELFKPMAVKAIRTEVLGGLGGFAGLFAL
MSMEG_5798|M.smegmatis_MC2_155 -SSDAISYASAGVDIEAGDRAVELFKPLAAKATRPEVRGGLGGFAGLFAL
Mflv_1636|M.gilvum_PYR-GCK ADQHGISYASAGVDIEAGDRAVELFKPLAKKATRPEVRGDLGGFAGLFAL
Mvan_5111|M.vanbaalenii_PYR-1 AEQHGISYASAGVDIEAGDRAVELFKPLAKKATRPEVRGGLGGFAGLFAL
TH_0070|M.thermoresistible__bu AEQHGISYASAGVDIEAGDRAVELIKPLASRASRPEVMGGLGGFAGLFKL
MAB_0728|M.abscessus_ATCC_1997 ADKPSNSYAAAGVDIEAGDRAVELFKKSVAKTHRPEVLGGIGGFAGLFAL
. . :**::*********:::*:* . :: *.** *.:******* *
MUL_0418|M.ulcerans_Agy99 RNDYREPVLAASTDGVGTKLAVAQAMDKHDTVGLDLVAMVVDDLVVCGAE
MMAR_4880|M.marinum_M RNDYREPVLAASTDGVGTKLAVAQAMDKHDTVGLDLVAMVVDDLVVCGAE
MAV_0748|M.avium_104 RGEYREPLLAASTDGVGTKLAVAQAMDKHDTVGLDLVAMVVDDLVVCGAE
Mb0832|M.bovis_AF2122/97 RGDYREPVLAASSDGVGTKLAIAQAMDKHDTVGLDLVAMVVDDLVVCGAE
Rv0809|M.tuberculosis_H37Rv RGDYREPVLAASSDGVGTKLAIAQAMDKHDTVGLDLVAMVVDDLVVCGAE
MLBr_02205|M.leprae_Br4923 RSGYREPVLAASTDGVGTKLAVAQAMDKHDTVGLDLVAMVVDDLVVCGAE
MSMEG_5798|M.smegmatis_MC2_155 KRDYREPVLASSTDGVGTKLAIAQAMDKHDTVGLDLVAMVVDDLVVCGAE
Mflv_1636|M.gilvum_PYR-GCK RGDYREPLLASSTDGVGTKLAVAQAMDKHDTVGIDLVAMVVDDLVVCGAE
Mvan_5111|M.vanbaalenii_PYR-1 RGDYREPLLASSTDGVGTKLAVAQAMDKHDTVGIDLVAMVVDDLVVCGAE
TH_0070|M.thermoresistible__bu RGGYREPILASSTDGVGTKLAVAQAMDKHDTVGLDLVAMVVDDLVVCGAE
MAB_0728|M.abscessus_ATCC_1997 KGGYREPVLAASTDGVGTKIAVAQAMDKRDTVGLDLVAMVVDDLVVCGAE
: ****:**:*:******:*:******:****:****************
MUL_0418|M.ulcerans_Agy99 PLFLQDYIAVGRTVPERVSAIVSGIAEGCVRAGCALLGGETAEHPGLMAP
MMAR_4880|M.marinum_M PLFLQDYIAVGRTVPERVSAIVSGIAEGCVRAGCALLGGETAEHPGLMAP
MAV_0748|M.avium_104 PLFLQDYIAVGRTVPERLSAIVSGIAEGCVRAGCALLGGETAEHPGLMEP
Mb0832|M.bovis_AF2122/97 PLFLLDYIAVGRIVPERLSAIVAGIADGCMRAGCALLGGETAEHPGLIEP
Rv0809|M.tuberculosis_H37Rv PLFLLDYIAVGRIVPERLSAIVAGIADGCMRAGCALLGGETAEHPGLIEP
MLBr_02205|M.leprae_Br4923 PLFLQDYIAVGRTVPERLSAIVDGIAEGCMLAGCALLGGETAEHPGLMEV
MSMEG_5798|M.smegmatis_MC2_155 PLFLQDYIAVGRTVPERVSAIVAGIAEGCVRAGCALLGGETAEHPGLMEP
Mflv_1636|M.gilvum_PYR-GCK PLFLQDYIAIGRTVPERVAELVSGIADGCVQAGCALLGGETAEHPGLMAP
Mvan_5111|M.vanbaalenii_PYR-1 PLFLQDYIAVGRTVPERVAELVSGIADGCVLAGCALLGGETAEHPGLMAP
TH_0070|M.thermoresistible__bu PLFLQDYIAVGRTVPERISELVSGIAEGCVQAGCALLGGETAEHPGLMAP
MAB_0728|M.abscessus_ATCC_1997 PLFLQDYIAVGKVVPERVSELVAGIAEGCAIAGCALLGGETAEHPGLMAP
**** ****:*: ****:: :* ***:** ****************:
MUL_0418|M.ulcerans_Agy99 DHYDISATGVGVVEADNVLGPERVKPGDVIIAMGSSGLHSNGYSLARNVL
MMAR_4880|M.marinum_M DHYDISATGVGVVEADDVLGPERVKPGDVIIAMGSSGLHSNGYSLARNVL
MAV_0748|M.avium_104 DHYDISATGVGVVEADDVLGPDRVKPGDVIIAMGSSGLHSNGYSLARTVL
Mb0832|M.bovis_AF2122/97 DHYDISATGVGVVEADNVLGPDRVKPGDVIIAMGSSGLHSNGYSLVRKVL
Rv0809|M.tuberculosis_H37Rv DHYDISATGVGVVEADNVLGPDRVKPGDVIIAMGSSGLHSNGYSLVRKVL
MLBr_02205|M.leprae_Br4923 DHYDIAATGVGVVEADDTLGPDRVKPGDVIIALRSSGLHSNGYSLARSVL
MSMEG_5798|M.smegmatis_MC2_155 DHYDISATGVGIVEADDVLGPDRVRPGDVIIAMASSGLHSNGYSLARKVL
Mflv_1636|M.gilvum_PYR-GCK DHYDISATGIGVVEADDVLGPDRVRPGDVIIAMASTGLHSNGYSLARHVL
Mvan_5111|M.vanbaalenii_PYR-1 DHYDISATGIGVVEADDVLGPDRVRPGDVIIGMASTGLHSNGYSLARHVL
TH_0070|M.thermoresistible__bu DHYDLSAFGVGIVEADDLLGPERVRPGDVIIAMGSSGLHSNGYSLARKVL
MAB_0728|M.abscessus_ATCC_1997 DDFDLSATGVGIVEADRVLGPDRVRPGDVVIAMGSSGLHSNGYSLARHVL
*.:*::* *:*:**** ***:**:****:*.: *:*********.* **
MUL_0418|M.ulcerans_Agy99 LEIDRMNLAGYVEEFGRTLGEELLEPTRIYAKDCLALTAETHVRTFCHVT
MMAR_4880|M.marinum_M LEIDRMNLAGYVEEFGRTLGEELLEPTRIYAKDCLALTAETHVRTFCHVT
MAV_0748|M.avium_104 LEIDRMNLEGYVEEFGRTLGEELLEPTRIYAKDCLALAAETHVRTFCHVT
Mb0832|M.bovis_AF2122/97 LEIDRMNLAGHVEEFGRTLGEELLEPTRIYAKDCLALAAETRVRTFCHVT
Rv0809|M.tuberculosis_H37Rv LEIDRMNLAGHVEEFGRTLGEELLEPTRIYAKDCLALAAETRVRTFCHVT
MLBr_02205|M.leprae_Br4923 LEIDRMNLAGYVEEFGRTLGEELLEPTRIYAKDCLALAAETHVRTFCHVT
MSMEG_5798|M.smegmatis_MC2_155 LEIDRMNLAGHVEEFGRTLGEELLEPTLIYAKDCLALAAETQVRTFCHVT
Mflv_1636|M.gilvum_PYR-GCK LEIDHMNLAGHVEEFGRTLGEELLEPTRIYAKDCLALAAETQVRTFCHVT
Mvan_5111|M.vanbaalenii_PYR-1 LEIDHMNLAGHVEEFGRTLGEELLEPTRIYAKDCLALAAETQVRTFCHVT
TH_0070|M.thermoresistible__bu LEIDRMNLAGHVEEFGRTLGEELLEPTRIYARDCLALAAETNVRTFCHIT
MAB_0728|M.abscessus_ATCC_1997 LEIDRMDLFGQVEEFGRTLGEELLEPTRIYAKDCLALAAETEVRTFCHVT
****:*:* * **************** ***:*****:***.******:*
MUL_0418|M.ulcerans_Agy99 GGGLAGNLERVIPHGLVAEIDRGTWTPAPVFAMIAQRGRVVREEMEKTFN
MMAR_4880|M.marinum_M GGGLAGNLERVIPHGLVAEIDRGTWTPAPVFAMIAQRGRVVREEMEKTFN
MAV_0748|M.avium_104 GGGLAGNLARVIPHGLVAELDRGTWTPAPVFAMIAQRGRVTREEMEKTFN
Mb0832|M.bovis_AF2122/97 GGGLAGNLQRVIPHGLIAEVDRGTWTPAPVFTMIAQRGRVRRTEMEKTFN
Rv0809|M.tuberculosis_H37Rv GGGLAGNLQRVIPHGLIAEVDRGTWTPAPVFTMIAQRGRVRRTEMEKTFN
MLBr_02205|M.leprae_Br4923 GGGLAGNLQRVIPHGLVAEVDRGTWTPAPVFAMIAQRGRVAREEMEKTFN
MSMEG_5798|M.smegmatis_MC2_155 GGGLAGNLERVIPHGLTAELDRGTWTPAPVFGMIAQRGRIERAEMEKTFN
Mflv_1636|M.gilvum_PYR-GCK GGGLAGNLERVIPHGLVAQLDRGTWTPAPVFGMIAQRGRIERAEMEKTFN
Mvan_5111|M.vanbaalenii_PYR-1 GGGLAGNLERVIPHGLVAELDRGTWTPAPVFGMIAQRGRIERAEMEKTFN
TH_0070|M.thermoresistible__bu GGGLAGNLERVIPNGLSAELDRGTWTPAPVFAMIAQRGRIERAEMERTFN
MAB_0728|M.abscessus_ATCC_1997 GGGLAGNLARVIPDGLVAELDRGSWTPGPIFAMIAQRGRIERAEMEKTFN
******** ****.** *::***:***.*:* *******: * ***:***
MUL_0418|M.ulcerans_Agy99 MGVGMIAVVAPEDTDRALAILTARHLDCWVLGTVCKGGKEG--PRATLVG
MMAR_4880|M.marinum_M MGVGMIAVVAPEDTDRALAILTARHLDCWVLGTVCKGGKEG--PRATLVG
MAV_0748|M.avium_104 MGVGMIAVVAPEDTDRALAILTARHLDCWVLGTVGKGGKAG--PRAKLVG
Mb0832|M.bovis_AF2122/97 MGVGMIAVVAPEDTTRALAVLTARHLDCWVLGTVCKGGKQG--PRAKLVG
Rv0809|M.tuberculosis_H37Rv MGVGMIAVVAPEDTTRALAVLTARHLDCWVLGTVCKGGKQG--PRAKLVG
MLBr_02205|M.leprae_Br4923 MGVGMIAVVAPEDTDRALAILTARHLDCWVLGTVCKGSKVG--PRAKLVG
MSMEG_5798|M.smegmatis_MC2_155 MGVGMVAVVAPEDTDRALAILTARHLNCWTLGTVKKGGKDS--VRAQLVG
Mflv_1636|M.gilvum_PYR-GCK MGVGMVAVVAPEDTDRALAVLTARHLDCWTLGTIEKGAKDG--SRAVLVG
Mvan_5111|M.vanbaalenii_PYR-1 MGVGMVAIVAPEDTDRALAVLTARHLNCWTLGTIEKGGKDG--PRAVLVG
TH_0070|M.thermoresistible__bu MGVGMVAIVAPEDTDRALAILTARHLDCWVLGTVTKAGKDE--PRARLVG
MAB_0728|M.abscessus_ATCC_1997 MGVGMVAVVAPEDVDRALAVLTARHIDCWTLGTIKKSGKDSGSESALLVG
*****:*:*****. ****:*****::**.***: *..* * ***
MUL_0418|M.ulcerans_Agy99 QHPRF
MMAR_4880|M.marinum_M QHPRF
MAV_0748|M.avium_104 QHPRF
Mb0832|M.bovis_AF2122/97 QHPRF
Rv0809|M.tuberculosis_H37Rv QHPRF
MLBr_02205|M.leprae_Br4923 QHPRF
MSMEG_5798|M.smegmatis_MC2_155 QHPRF
Mflv_1636|M.gilvum_PYR-GCK QHPRF
Mvan_5111|M.vanbaalenii_PYR-1 QHPRF
TH_0070|M.thermoresistible__bu QHPRF
MAB_0728|M.abscessus_ATCC_1997 QHPRF
*****