For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTARIGVITFPGTLDDVDAARAIRLVGAEAVNLWHADADLKGVEAVVVPGGFSYGDYLRAGAIARFAPVM GEVVSAARRGMPVLGICNGFQVLCEAGLLPGALTRNVGLHFVCRDAWLRVASTSTAWTSRFEPAADLLVP LKSGEGRYVASEQVLDELEAEDRVVFRYHDNVNGSLRDIAGISSANGRVVGLMPHPEHAIEALTGPSDDG LGLFYSALDAVLAA
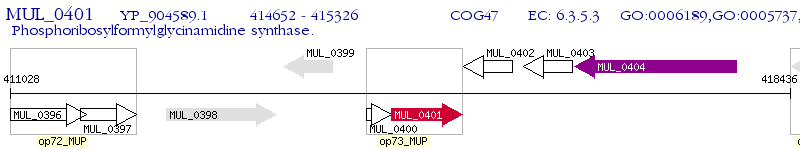
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0401 | purQ | - | 100% (224) | phosphoribosylformylglycinamidine synthase I |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0812 | purQ | 1e-117 | 92.34% (222) | phosphoribosylformylglycinamidine synthase I |
| M. gilvum PYR-GCK | Mflv_1625 | - | 1e-111 | 85.71% (224) | phosphoribosylformylglycinamidine synthase subunit I |
| M. tuberculosis H37Rv | Rv0788 | purQ | 1e-117 | 92.34% (222) | phosphoribosylformylglycinamidine synthase I |
| M. leprae Br4923 | MLBr_02219 | purQ | 1e-113 | 87.50% (224) | phosphoribosylformylglycinamidine synthase I |
| M. abscessus ATCC 19977 | MAB_0698 | - | 1e-106 | 82.59% (224) | phosphoribosylformylglycinamidine synthase I |
| M. marinum M | MMAR_4897 | purQ | 1e-125 | 99.11% (224) | phosphoribosylformylglycinamidine synthase I, PurQ |
| M. avium 104 | MAV_0736 | purQ | 1e-114 | 89.24% (223) | phosphoribosylformylglycinamidine synthase subunit I |
| M. smegmatis MC2 155 | MSMEG_5831 | purQ | 1e-109 | 82.96% (223) | phosphoribosylformylglycinamidine synthase subunit I |
| M. thermoresistible (build 8) | TH_0567 | purQ | 1e-108 | 82.14% (224) | PROBABLE PHOSPHORIBOSYLFORMYLGLYCINAMIDINE SYNTHASE I PURG |
| M. vanbaalenii PYR-1 | Mvan_5128 | - | 1e-111 | 84.38% (224) | phosphoribosylformylglycinamidine synthase subunit I |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0401|M.ulcerans_Agy99 MTARIGVITFPGTLDDVDAARAIRLVGAEAVNLWHADADLKGVEAVVVPG
MMAR_4897|M.marinum_M MTARIGVITFPGTLDDVDAARAIRLVGAEAVNLWHADADLKGVEAVVVPG
Mb0812|M.bovis_AF2122/97 MTARIGVVTFPGTLDDVDAARAARQVGAEVVSLWHADADLKGVDAVVVPG
Rv0788|M.tuberculosis_H37Rv MTARIGVVTFPGTLDDVDAARAARQVGAEVVSLWHADADLKGVDAVVVPG
MAV_0736|M.avium_104 MTARIGIITFPGTLDDVDAARAARRVGAQPVSLWHADADLKGVDAVVVPG
MLBr_02219|M.leprae_Br4923 MNARIGVITFPGTLDDVDAARAARHVGAEAVSLWHADADLKGVDAVVVPG
Mflv_1625|M.gilvum_PYR-GCK MTARVGVITFPGTLDDIDAARAVRLAGGEAVNLWHADADLKGVDAVIVPG
Mvan_5128|M.vanbaalenii_PYR-1 MTARVGVITFPGTLDDIDAARAVRLAGAEAVSLWHADADLKGVDAVVVPG
MSMEG_5831|M.smegmatis_MC2_155 MTTRVGVITFPGTLDDIDAARAVRLAGAEAVSLWHADADLHGVDAVVVPG
TH_0567|M.thermoresistible__bu MSARIGVITFPGTLDDVDAARAVRLAGAEPVTLWHGDADLKGVDAVIVPG
MAB_0698|M.abscessus_ATCC_1997 MTARIGVITFPGTLDDVDAARAARLSGAEAVSLWHDDADLKSVDAVIVPG
*.:*:*::********:***** * *.: *.*** ****:.*:**:***
MUL_0401|M.ulcerans_Agy99 GFSYGDYLRAGAIARFAPVMGEVVSAARRGMPVLGICNGFQVLCEAGLLP
MMAR_4897|M.marinum_M GFSYGDYLRAGAIARFAPVMGEVVSAAQRGMPVLGICNGFQVLCEAGLLP
Mb0812|M.bovis_AF2122/97 GFSYGDYLRAGAIARFAPVMDEVVAAADRGMPVLGICNGFQVLCEAGLLP
Rv0788|M.tuberculosis_H37Rv GFSYGDYLRAGAIARFAPVMDEVVAAADRGMPVLGICNGFQVLCEAGLLP
MAV_0736|M.avium_104 GFSYGDYLRAGAIARFAPVMTEVVDAARRGMPVLGICNGFQVLCEAGLLP
MLBr_02219|M.leprae_Br4923 GFSYGDYLRAGAIARLSPIMTEVVDAVQRGMPVLGICNGFQVLCEAGLLP
Mflv_1625|M.gilvum_PYR-GCK GFSYGDYLRAGAIARFAPVMGEVVAAAGRGMPVLGICNGFQVLCEAGLLP
Mvan_5128|M.vanbaalenii_PYR-1 GFSYGDYLRAGAIAKFAPVMGEVIEAAGRGMPVLGICNGFQVLCEAGLLP
MSMEG_5831|M.smegmatis_MC2_155 GFSYGDYLRCGAIAKFAPVMGSVVEAANKGMPVLGICNGFQVLCEAGLLP
TH_0567|M.thermoresistible__bu GFSYGDYLRAGAIAKFAPVMGEVIEQAGRGLPVLGICNGFQVLCEAGLLP
MAB_0698|M.abscessus_ATCC_1997 GFSYGDYLRAGAIARFAPVMRSVVDAAAQGLPILGICNGFQVLCEAGLLP
*********.****:::*:* .*: . :*:*:*****************
MUL_0401|M.ulcerans_Agy99 GALTRNVGLHFVCRDAWLRVASTSTAWTSRFEPAADLLVPLKSGEGRYVA
MMAR_4897|M.marinum_M GALTRNVGLHFVCRDAWLRVASTSTAWTSRFEPDADLLVPLKSGEGRYVA
Mb0812|M.bovis_AF2122/97 GALTRNVGLHFICRDVWLRVASTSTAWTSRFEPDADLLVPLKSGEGRYVA
Rv0788|M.tuberculosis_H37Rv GALTRNVGLHFICRDVWLRVASTSTAWTSRFEPDADLLVPLKSGEGRYVA
MAV_0736|M.avium_104 GALTRNVGLHFICRDVWLRVASTSTAWTSRFEPDADLLVPLKSGEGRYVA
MLBr_02219|M.leprae_Br4923 GALIRNVGLHFICRDVWLRVISTSTAWTSRFEPETDLLVSLKSGEGRYVA
Mflv_1625|M.gilvum_PYR-GCK GALTRNENLHFICRDTWLEVASNSSAWTSRYEQGADLLVPLKSGEGRYVA
Mvan_5128|M.vanbaalenii_PYR-1 GALTRNAGLHFICRDTWLEVASNTTAWTTRYEQGADLLVPLKSGEGRYVA
MSMEG_5831|M.smegmatis_MC2_155 GALTRNAGLHFVCRDVWLEVASNTSAWTTRYETGADLLIPLKSGEGRYVA
TH_0567|M.thermoresistible__bu GALTRNIGLHFICRDVWLEVASNSTAWTTRYDVGADLLIPLKSGEGRYVA
MAB_0698|M.abscessus_ATCC_1997 GALTRNAGLHFVCRDVWLGVDANNTAWSSRYERDADILVPLKSGEGRYVA
*** ** .***:***.** * :..:**::*:: :*:*:.**********
MUL_0401|M.ulcerans_Agy99 SEQVLDELEAEDRVVFRYHDNVNGSLRDIAGISSANGRVVGLMPHPEHAI
MMAR_4897|M.marinum_M SEQVLDELEAEDRVVFRYHDNVNGSLRDIAGISSANGRVVGLMPHPEHAI
Mb0812|M.bovis_AF2122/97 PEKVLDELEGEGRVVFRYHDNVNGSLRDIAGICSANGRVVGLMPHPEHAI
Rv0788|M.tuberculosis_H37Rv PEKVLDELEGEGRVVFRYHDNVNGSLRDIAGICSANGRVVGLMPHPEHAI
MAV_0736|M.avium_104 PADVLDELEGEGRVVFRYHENLNGSQRDIAGVSSADGRVVGLMPHPEHAI
MLBr_02219|M.leprae_Br4923 SENVLDELDGEGRVVFRYHDNINGSLRDIAGISSANGRVVGMMPHPEHAI
Mflv_1625|M.gilvum_PYR-GCK STEVLDELEGEGRVVFRYRENLNGSMRAIAGISSANGRVVGLMPHPEHAT
Mvan_5128|M.vanbaalenii_PYR-1 GEAVLDELEGEGRVVFRYRENLNGSMRGIAGVSSANGRVVGLMPHPEHAT
MSMEG_5831|M.smegmatis_MC2_155 SESVLDELEGEDRVVFRYRENLNGSMRDIAGICSENRRVVGLMPHPEHAT
TH_0567|M.thermoresistible__bu PESVLDELEGEGRVVFRYRENLNGSQRDIAGISSANGRVVGLMPHPEHAT
MAB_0698|M.abscessus_ATCC_1997 SDDVLDELEGEGRVVFRYRENPNGSMRDIAGISSANGRVVGLMPHPEHAT
*****:.*.******::* *** * ***:.* : ****:*******
MUL_0401|M.ulcerans_Agy99 EALTGPSDDGLGLFYSALDAVLAA-
MMAR_4897|M.marinum_M EALTGPSDDGLGLFYSALDAVLAA-
Mb0812|M.bovis_AF2122/97 EALTGPSDDGLGLFYSALDAVLTG-
Rv0788|M.tuberculosis_H37Rv EALTGPSDDGLGLFYSALDAVLTG-
MAV_0736|M.avium_104 EALTGPSDDGLGLFYSVLDGVLAG-
MLBr_02219|M.leprae_Br4923 EVLTGPSDDGLGLFYSALDSVLAS-
Mflv_1625|M.gilvum_PYR-GCK EALTGPSDDGLGIFYSALDAVLVA-
Mvan_5128|M.vanbaalenii_PYR-1 EALTGPSDDGLGIFYSALDAVLVS-
MSMEG_5831|M.smegmatis_MC2_155 EALTGPSDDGLGLFYSALDAILSV-
TH_0567|M.thermoresistible__bu EALTGPSDDGLGMFLSVLDSVLTAA
MAB_0698|M.abscessus_ATCC_1997 EPLTGPSDDGLGIFYSALDAVLTAG
* **********:* *.**.:*