For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVTALGVPTVRGRSEGSAVASDAPGDGPLLDRFGRSATDLRVSLTDRCNLRCGYCMPAEGLNWLPGEQL LGPAELARLPRIAVTPLGITSVRFTGGEPLLARHLEEVVAAAAQLRPRPEISLTTNGVGLAKRAAALAEA GLDRVNVSLDTVDRAHFAAVTRRDRLTDVLDGLAGARAAGLTPVKVNAVLDPETGRQDVVELLRFCLEQG YQLRVIEQMPLDAGHQWRRNALLGSDDVLAALQPHFRLRPDPAPRGSAPAELWLVDAGPDTPAGKFGVIA SVSHAFCSTCDRTRLTADGQVRSCLFSTEETDLRGLLRAGAGDEAIEAAWRGAMWAKPAGHGINNPDFLQ PQRPMSAIGG
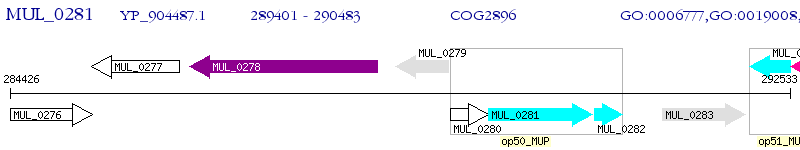
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0281 | moaA | - | 100% (360) | molybdenum cofactor biosynthesis protein A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0893c | moaA | 1e-172 | 81.67% (360) | molybdenum cofactor biosynthesis protein A |
| M. gilvum PYR-GCK | Mflv_1705 | moaA | 1e-142 | 68.61% (360) | molybdenum cofactor biosynthesis protein A |
| M. tuberculosis H37Rv | Rv0869c | moaA | 1e-172 | 81.67% (360) | molybdenum cofactor biosynthesis protein A |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0871c | moaA | 1e-125 | 66.27% (332) | molybdenum cofactor biosynthesis protein A |
| M. marinum M | MMAR_4663 | moaA | 0.0 | 98.33% (360) | molybdenum cofactor biosynthesis protein A2 MoaA2 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5698 | moaA | 1e-155 | 74.79% (361) | molybdenum cofactor biosynthesis protein A |
| M. thermoresistible (build 8) | TH_1071 | moaA | 1e-152 | 74.72% (360) | molybdenum cofactor biosynthesis protein A |
| M. vanbaalenii PYR-1 | Mvan_5047 | moaA | 1e-141 | 70.64% (361) | molybdenum cofactor biosynthesis protein A |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1705|M.gilvum_PYR-GCK MTVVPLGLPSVQRPS-------TPAPATGPLIDTFGRVATDLRVSLTDLC
Mvan_5047|M.vanbaalenii_PYR-1 MTVVALGVPSVHRPQ-------SPAPEEGPLVDTFGRVATDLRVSLTDLC
MSMEG_5698|M.smegmatis_MC2_155 MTVTALGLPTVARSTGDGSAGASPAPADGPLVDTYGRAATDLRVSLTDRC
MUL_0281|M.ulcerans_Agy99 MTVTALGVPTVRGRSEG-SAVASDAPGDGPLLDRFGRSATDLRVSLTDRC
MMAR_4663|M.marinum_M MTVTALGVPTVRGRSEG-SAIASDAPGDGPLLDRFGRSATDLRVSLTDRC
Mb0893c|M.bovis_AF2122/97 MTLTALGMPALRSRTNG-IADPRVVPTTGPLVDTFGRVANDLRVSLTDRC
Rv0869c|M.tuberculosis_H37Rv MTLTALGMPALRSRTNG-IADPRVVPTTGPLVDTFGRVANDLRVSLTDRC
TH_1071|M.thermoresistible__bu MTVTALGLPAVRRPSP-----PSAPPASGPLVDTFGRVHTDLRISLTDRC
MAB_0871c|M.abscessus_ATCC_199 ---------------------------MTPLIDSFGRIATDLRVSLTDRC
**:* :** .***:**** *
Mflv_1705|M.gilvum_PYR-GCK NLRCTYCMPAEGLDWMPRDQKLTPDELTRLLRIGVTRLGITSIRFTGGEP
Mvan_5047|M.vanbaalenii_PYR-1 NLRCTYCMPAEGLDWLPSDEKLNPDELIRLLGVAVTRLGITSVRFTGGEP
MSMEG_5698|M.smegmatis_MC2_155 NLRCTYCMPAEGLNWLPGDALLSPTELARLLRIAVARLGITSVRFTGGEP
MUL_0281|M.ulcerans_Agy99 NLRCGYCMPAEGLNWLPGEQLLGPAELARLPRIAVTPLGITSVRFTGGEP
MMAR_4663|M.marinum_M NLRCGYCMPAEGLNWLPGEQLLGPAELARLLRIAVTRLGITSVRFTGGEP
Mb0893c|M.bovis_AF2122/97 NLRCSYCMPERGLRWLPGEQLLRPDELARLIHIAVTRLGVTSVRFTGGEP
Rv0869c|M.tuberculosis_H37Rv NLRCSYCMPERGLRWLPGEQLLRPDELARLIHIAVTRLGVTSVRFTGGEP
TH_1071|M.thermoresistible__bu NLRCTYCMPAEGLDWLPGEHLLRPDELARLLHIAVTRLGITNVRFTGGEP
MAB_0871c|M.abscessus_ATCC_199 NLRCTYCMPAEGLDWLRSETLLTTDEYIRLIGIAVTRLGIDSVRFTGGEP
**** **** .** *: : * . * ** :.*: **: .:*******
Mflv_1705|M.gilvum_PYR-GCK LVVPHLEDVLAATTALQPRPEITLTTNGIGLAQRAAGLRRAGLNRINVSL
Mvan_5047|M.vanbaalenii_PYR-1 LVVPHLEDVVAATAALRPRPEITLTTNGVGLAKRAAGLKRAGLDRINVSL
MSMEG_5698|M.smegmatis_MC2_155 LVVRHLEEVVAAAAALEPRPEITLTTNGLGLARRAEGLKKAGLNRINVSL
MUL_0281|M.ulcerans_Agy99 LLARHLEEVVAAAAQLRPRPEISLTTNGVGLAKRAAALAEAGLDRVNVSL
MMAR_4663|M.marinum_M LLARHLEEVVAAAAQLRPRPEISLTTNGVGLAKRAAALAEAGLDRVNVSL
Mb0893c|M.bovis_AF2122/97 LLAHHLDEVVAATARLRPRPEISLTTNGVGLARRAGALAEAGLDRVNVSL
Rv0869c|M.tuberculosis_H37Rv LLAHHLDEVVAATARLRPRPEISLTTNGVGLARRAGALAEAGLDRVNVSL
TH_1071|M.thermoresistible__bu LVARHLEEVVAAAAAERPRPEIALTTNGIGLAKRARRLKDAGLDRVNVSL
MAB_0871c|M.abscessus_ATCC_199 LIFKDLEKLVAAVAGMSPRPDIAVTTNGVGLDRKAVALREAGLDRINVSL
*: .*:.::**.: ***:*::****:** ::* * ***:*:****
Mflv_1705|M.gilvum_PYR-GCK DTVDPARFAAITRRDRFDDVIAGLHAAKEAGLQPVKVNAVLDPVTGLQDA
Mvan_5047|M.vanbaalenii_PYR-1 DTVDPARFAAITRRDRFTEVVAGLRAAKEAGLDPVKVNAVLDPVSGLDDV
MSMEG_5698|M.smegmatis_MC2_155 DSVDAAHFARITRRDRLPDVLAGLAAAKAAGLEPVKVNAVLDPVSGLDDA
MUL_0281|M.ulcerans_Agy99 DTVDRAHFAAVTRRDRLTDVLDGLAGARAAGLTPVKVNAVLDPETGRQDV
MMAR_4663|M.marinum_M DTVDRAHFAAITRRDRLTDVLDGLAGARAAGLTPVKVNAVLDPETGRQDV
Mb0893c|M.bovis_AF2122/97 DSIDRAHFAAITRRDRLAHVLAGLAAAKAAGLTPVKVNAVLDPTTGREDV
Rv0869c|M.tuberculosis_H37Rv DSIDRAHFAAITRRDRLAHVLAGLAAAKAAGLTPVKVNAVLDPTTGREDV
TH_1071|M.thermoresistible__bu DTVDPVRFARITRRDRLADVLAGLEAARAAGLTPVKVNAVLDAETGLDDA
MAB_0871c|M.abscessus_ATCC_199 DTLDAHRFAAITRRDRLSDVLLGLKAAGSAGLSPIKINAVLTPEQYKEDA
*::* :** :*****: .*: ** .* *** *:*:**** . :*.
Mflv_1705|M.gilvum_PYR-GCK VALLGFCLENGYQLRIIEQMPLDAGHSWQRDRALSADDILDTLRRHFTLT
Mvan_5047|M.vanbaalenii_PYR-1 VSLLRFCLEHGYQLRVIEQMPLDAGHSWERERALSADVILSALRRHFTLT
MSMEG_5698|M.smegmatis_MC2_155 VELLRYCLQHGYQLRIIEQMPLDASHSWQRDNVIDADRILQTLQQHFELT
MUL_0281|M.ulcerans_Agy99 VELLRFCLEQGYQLRVIEQMPLDAGHQWRRNALLGSDDVLAALQPHFRLR
MMAR_4663|M.marinum_M VELLRFCLEQGYQLRVIEQMPLDAGHQWRRNALLGSDDVLAALQPHFRLR
Mb0893c|M.bovis_AF2122/97 VDLLRFCLERGYQLRVIEQMPLDAGHSWRRNIALSADDVLAALRPHFRLR
Rv0869c|M.tuberculosis_H37Rv VDLLRFCLERGYQLRVIEQMPLDAGHSWRRNIALSADDVLAALRPHFRLR
TH_1071|M.thermoresistible__bu VQLLRFCLDRGYQLRIIEQMPLDAGHQWRRDQTLSADEVLAALQREFRLS
MAB_0871c|M.abscessus_ATCC_199 VALLHFSLEHGYELRIIEQMPLDAGHSWSRARMVTADEIHAALSREFSLS
* ** :.*:.**:**:********.*.* * : :* : :* .* *
Mflv_1705|M.gilvum_PYR-GCK PDPAPRGSAPAQLWRVHDG-TGVVGRVGIIASVSHAFCSTCDRSRLTADG
Mvan_5047|M.vanbaalenii_PYR-1 PDPRPRGSAPAQLWQVHDD-GHVTGHIGIIASVSDAFCSTCDRSRLTADG
MSMEG_5698|M.smegmatis_MC2_155 PDSRPRGSAPAELWQVAEGPTHAAGTVGVIASVSHAFCSACDRTRLTADG
MUL_0281|M.ulcerans_Agy99 PDPAPRGSAPAELWLVDAGPDTPAGKFGVIASVSHAFCSTCDRTRLTADG
MMAR_4663|M.marinum_M PDPAPRGSAPAELWLVDAGPDTPAGKFGVIASVSHAFCSTCDRTRLTADG
Mb0893c|M.bovis_AF2122/97 PDPAPRGSAPAELWLVDAGPNTPRGRFGVIASVSHAFCSTCDRTRLTADG
Rv0869c|M.tuberculosis_H37Rv PDPAPRGSAPAELWLVDAGPNTPRGRFGVIASVSHAFCSTCDRTRLTADG
TH_1071|M.thermoresistible__bu PDPTPRGSAPAQLWRIDG----TGATVGIIATVSHAFCSACDRTRLTADG
MAB_0871c|M.abscessus_ATCC_199 EDPAPRGSAPAQRWLVNGG----PATVGIIASVSRPFCGDCDRTRLTADG
*. *******: * : . .*:**:** .**. ***:******
Mflv_1705|M.gilvum_PYR-GCK QVRNCLFARDETDLRALLRSGADDDVLEAAWRAAMWAKAAGHGINDPDFV
Mvan_5047|M.vanbaalenii_PYR-1 QVRNCLFARDETDLRALLRSGADDAALEDAWRAAMWAKAAGHGINDPGFV
MSMEG_5698|M.smegmatis_MC2_155 QIRSCLFSREETDLRHLLRGGADDAEIEAAWRAAMWSKPAGHGINDPDFV
MUL_0281|M.ulcerans_Agy99 QVRSCLFSTEETDLRGLLRAGAGDEAIEAAWRGAMWAKPAGHGINNPDFL
MMAR_4663|M.marinum_M QVRSCLFSTEETDLRGLLRAGAGDEAIEAAWRSAMWAKPAGHGINNPDFV
Mb0893c|M.bovis_AF2122/97 QIRSCLFSTEETDLRRLLRGGADDDAIEAAWRAAMWSKPAGHGINAPDFI
Rv0869c|M.tuberculosis_H37Rv QIRSCLFSTEETDLRRLLRGGADDDAIEAAWRAAMWSKPAGHGINAPDFI
TH_1071|M.thermoresistible__bu QVRNCLFAQQETDLRGLLRSGADDDAVEAAWRAAMWAKAAGHGINDPDFV
MAB_0871c|M.abscessus_ATCC_199 QVRNCLFATTETDLRGLLRSGASDAEVASAWQAAMWGKKPGHGINDPSFL
*:*.***: ***** ***.**.* : **:.***.* .***** *.*:
Mflv_1705|M.gilvum_PYR-GCK QPDRPMSAIGG
Mvan_5047|M.vanbaalenii_PYR-1 QPDRPMSAIGG
MSMEG_5698|M.smegmatis_MC2_155 QPVRPMSAIGG
MUL_0281|M.ulcerans_Agy99 QPQRPMSAIGG
MMAR_4663|M.marinum_M QPQRPMSAIGG
Mb0893c|M.bovis_AF2122/97 QPDRPMSAIGG
Rv0869c|M.tuberculosis_H37Rv QPDRPMSAIGG
TH_1071|M.thermoresistible__bu QPARPMSAIGG
MAB_0871c|M.abscessus_ATCC_199 QPDRPMSAIGG
** ********