For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDDLIRIPAQTAVLGSEAHYPEESPLRQVDVNAFRIQAHQVTNTQFAEFVDATGYLTVAERPLDPADYPG APPENLQPGSMVFTRTAGPVDLRHLSLWWTWTPGASWRHPAGPHSSIAKRADHPVVHVAYEDAHAYAGWA GLALPSEAEWEVAARGGLTGAAYTWGEESERPGQRLANFWHGDFPWRPEPGYGRTSPVGSFEPNGYGLFD MAGNVWEWTVDWYIDTPSARPAAEADSYDPQQPQFRVPRKVVKGGSFLCADSYCRRYRPAARRPQPIDTG VSHIGFRCVGRD*
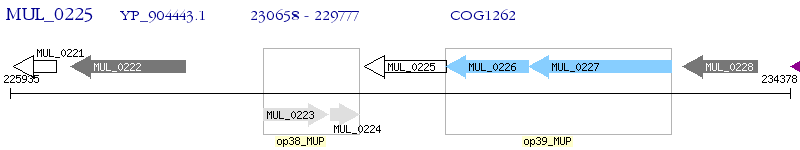
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0225 | - | - | 100% (293) | hypothetical protein MUL_0225 |
| M. ulcerans Agy99 | MUL_4287 | - | 5e-19 | 31.18% (263) | hypothetical protein MUL_4287 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0733 | - | 1e-88 | 55.00% (300) | hypothetical protein Mb0733 |
| M. gilvum PYR-GCK | Mflv_5038 | - | 2e-86 | 54.17% (288) | hypothetical protein Mflv_5038 |
| M. tuberculosis H37Rv | Rv0712 | - | 1e-88 | 55.00% (300) | hypothetical protein Rv0712 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4305 | - | 4e-84 | 53.26% (291) | hypothetical protein MAB_4305 |
| M. marinum M | MMAR_4408 | - | 0.0 | 99.32% (293) | hypothetical protein MMAR_4408 |
| M. avium 104 | MAV_5033 | - | 5e-91 | 55.86% (290) | sulfatase-modifying factor 1 |
| M. smegmatis MC2 155 | MSMEG_1226 | - | 1e-86 | 53.92% (293) | sulfatase-modifying factor 1 |
| M. thermoresistible (build 8) | TH_0314 | - | 1e-84 | 53.00% (300) | CONSERVED HYPOTHETICAL PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_4639 | - | 1e-137 | 74.65% (288) | hypothetical protein Mvan_4639 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_5033|M.avium_104 ------------------MLTE---LIDLPGGAFLMGSNSFYPEESPVHE
MSMEG_1226|M.smegmatis_MC2_155 ------------------MLTE---LVALDGGTFRMGSQDFYPEEAPVHE
MAB_4305|M.abscessus_ATCC_1997 MEYSPRNRKLREMETAAVTLTE---LVELPGGSFPMGCNVFYPEEMPERV
Mb0733|M.bovis_AF2122/97 ------------------MLTE---LVDLPGGSFRMGSTRFYPEEAPIHT
Rv0712|M.tuberculosis_H37Rv ------------------MLTE---LVDLPGGSFRMGSTRFYPEEAPIHT
Mflv_5038|M.gilvum_PYR-GCK ------------------MLTD---LVDLEGGSFRMGSTRFYPEEAPAHT
TH_0314|M.thermoresistible__bu ----------------MSALTD---LVEIPGGTFRMGSIRFYPEEAPVHT
MUL_0225|M.ulcerans_Agy99 ------------------MTDD---LIRIPAQTAVLGSEAHYPEESPLRQ
MMAR_4408|M.marinum_M ------------------MTDD---LIRIPAQTAVLGSEAHYPEESPLRQ
Mvan_4639|M.vanbaalenii_PYR-1 ------------------MTEHGAAMVWIPGQTAILGSDSHYPEEAPAHP
. :: : . : :*. .**** * :
MAV_5033|M.avium_104 VTVRPFSIERHPVTNAQFAEFVDQTGYVTVAEKALDPKDFPGVPAEDLKP
MSMEG_1226|M.smegmatis_MC2_155 VSVAPFSIERHPVTNAQFAEFVAATGYVTVAEQELDPAAFPGVPADELVP
MAB_4305|M.abscessus_ATCC_1997 SSVEPFAIERHPVTNAQFAEFVTQTGYVTVAERAPDPDMYPGASPDDLVP
Mb0733|M.bovis_AF2122/97 VTVRAFAVERHPVTNAQFAEFVSATGYVTVAEQPLDPGLYPGVDAADLCP
Rv0712|M.tuberculosis_H37Rv VTVRAFAVERHPVTNAQFAEFVSATGYVTVAEQPLDPGLYPGVDAADLCP
Mflv_5038|M.gilvum_PYR-GCK VTVAPFAIERNPVTNAQFAEFVDDTGYRTVAERPPDPALYPGASPHDLVP
TH_0314|M.thermoresistible__bu VTVADFAIERHPVTNAQFAEFVAATGYRTVAEQPLDPRLYPGVAAADLQP
MUL_0225|M.ulcerans_Agy99 VDVNAFRIQAHQVTNTQFAEFVDATGYLTVAERPLDPADYPGAPPENLQP
MMAR_4408|M.marinum_M VDVNAFRIQAHQVTNTQFAEFVDATGYLTVAERPLDPADYPGAPPENLQP
Mvan_4639|M.vanbaalenii_PYR-1 VTVDGFWMQPTQVTNLEFAEFVAATGYVTAAERPVNPADHPGAPRENLQP
* * :: *** :***** *** *.**: :* .**. :* *
MAV_5033|M.avium_104 GALVFTPTAGPVDLRAWRQWWTWMPGACWRQPFGPG-SSIEDRLDHPVVQ
MSMEG_1226|M.smegmatis_MC2_155 GALVFQPTEGPVNLRDWRQWWTWVPGACWKHPKGPG-SSIDDVPDHPVVQ
MAB_4305|M.abscessus_ATCC_1997 GAMTFRTTAGPVDLHDWQQWWDWVPGAYWRHPFGPG-SDIDGAAEHPVVQ
Mb0733|M.bovis_AF2122/97 GAMVFCPTAGPVDLRDWRQWWDWVPGACWRHPFGRD-SDIADRAGHPVVQ
Rv0712|M.tuberculosis_H37Rv GAMVFCPTAGPVDLRDWRQWWDWVPGACWRHPFGRD-SDIADRAGHPVVQ
Mflv_5038|M.gilvum_PYR-GCK GALVFRPTAGPVDLRDWRQWWEWTPGADWRHPFGPD-SSVDDRPEHPVVQ
TH_0314|M.thermoresistible__bu GALVFTPTRGPVDLRDWRQWWTWTPGACWRHPFGPEGPSWQDRPDHPVVQ
MUL_0225|M.ulcerans_Agy99 GSMVFTRTAGPVDLRHLSLWWTWTPGASWRHPAGPH-SSIAKRADHPVVH
MMAR_4408|M.marinum_M GSMVFTRTAGPVDLRHLSLWWTWTPGASWRHPAGPH-SSIAKRADHPVVH
Mvan_4639|M.vanbaalenii_PYR-1 GSMVFTRTGGPVDLRHLNLWWTWTPGASWRHPVGPA-SSIDKRLDHPVVH
*::.* * ***:*: ** * *** *::* * .. ****:
MAV_5033|M.avium_104 VAYPDAAAYASWAGRRLPSEAQWEYAARAGTS-SAYAWGDDVAPGGEIMA
MSMEG_1226|M.smegmatis_MC2_155 VAYPDAAAYAAWAGRRLPTEAEWEYAARAGAT-TVYAWGDDVRPDGQLMA
MAB_4305|M.abscessus_ATCC_1997 VAYTDAAAYARWAGRRLPTEAEWEYAARGGSG-TVYAWGDEVAPEGRLMA
Mb0733|M.bovis_AF2122/97 VAYPDAVAYARWAGRRLPTEAEWEYAARGGTT-ATYAWGDQEKPGGMLMA
Rv0712|M.tuberculosis_H37Rv VAYPDAVAYARWAGRRLPTEAEWEYAARGGTT-ATYAWGDQEKPGGMLMA
Mflv_5038|M.gilvum_PYR-GCK VCYSDAAAYARWAGRRLPTEAEWEFAAGAGST-AVYAWGDEPAPGGQLMA
TH_0314|M.thermoresistible__bu IAYPDAQAYARWAGRRLPTEREWEYAARAGAD-TVYPWGDEARPGGRLMA
MUL_0225|M.ulcerans_Agy99 VAYEDAHAYAGWAGLALPSEAEWEVAARGGLTGAAYTWGEESERPGQRLA
MMAR_4408|M.marinum_M VAYEDAHAYAGWAGLALPSEAEWEVAARGGLTGAAYTWGEESERPGQRLA
Mvan_4639|M.vanbaalenii_PYR-1 VGYEDAESYAAWAGLELPTEAEWETAARGGLTAATYTWGDVPEQPGRQLA
: * ** :** *** **:* :** ** .* :.*.**: * :*
MAV_5033|M.avium_104 NTWQGKFPYRNDGARGWTGTSPVGTFAPNGFGLVDMIGNVWEWTSTRYSA
MSMEG_1226|M.smegmatis_MC2_155 NTWQGRFPYRNDGALGWTGTSPVGTFPPNGFGLVDMIGNVWEWTSTRYTP
MAB_4305|M.abscessus_ATCC_1997 NTWQGRFPYRNDGALGWHGTSPVGTFPANDFGLLDMIGNVWEWTSTRFQH
Mb0733|M.bovis_AF2122/97 NTWQGRFPYRNDGALGWVGTSPVGRFPANGFGLLDMIGNVWEWTTTEFYP
Rv0712|M.tuberculosis_H37Rv NTWQGRFPYRNDGALGWVGTSPVGRFPANGFGLLDMIGNVWEWTTTEFYP
Mflv_5038|M.gilvum_PYR-GCK NTWQGAFPYRNDGALGWAGTSPVGTFPANGFGLVDMIGNVWEWTTTRFAG
TH_0314|M.thermoresistible__bu NTWQGRFPYRNDGAAGWIGTSPVGAFPPNDFGLFDMIGNVWEWTTTRFSG
MUL_0225|M.ulcerans_Agy99 NFWHGDFPWRPE--PGYGRTSPVGSFEPNGYGLFDMAGNVWEWTVDWYID
MMAR_4408|M.marinum_M NFWHGDFPWRPE--PGYGRTSPVGSFEPNGYGLFDMAGNVWEWTVDWYTD
Mvan_4639|M.vanbaalenii_PYR-1 NYWHGDFPWRPD--RGYGRTRPVGSFPPNRYGLFDMAGNVWEWTTDWYGD
* *:* **:* : *: * *** * .* :**.** ******* :
MAV_5033|M.avium_104 GHHP---GRAAPSCCPTSPT---GDPAVL-QTLKGGSHLCAPEYCHRYRP
MSMEG_1226|M.smegmatis_MC2_155 RHSR---RPEVSGCCP-APG---GDPSIH-QTLKGGSHLCAPEYCHRYRP
MAB_4305|M.abscessus_ATCC_1997 GAG------APKSCCSPSDN---PDPSVI-QVLKGGSHLCAPEYCHRYRP
Mb0733|M.bovis_AF2122/97 HHR---IDPPSTACCAPVKLATAADPTIS-QTLKGGSHLCAPEYCHRYRP
Rv0712|M.tuberculosis_H37Rv HHR---IDPPSTACCAPVKLATAADPTIS-QTLKGGSHLCAPEYCHRYRP
Mflv_5038|M.gilvum_PYR-GCK HHR--VNPPPPPTCCPPAD----PDPAVN-QALKGGSHLCAPEYCHRYRP
TH_0314|M.thermoresistible__bu HHRPAEADEPVRSCCTPSAPVERPDPAVS-QTLKGGSHLCAPEYCHRYRA
MUL_0225|M.ulcerans_Agy99 TPS------ARPAAEADSYDPQQPQFRVPRKVVKGGSFLCADSYCRRYRP
MMAR_4408|M.marinum_M TPS------ARPAAEADSYDPQQPQFRVPRKVVKGGSFLCADSYCRRYRP
Mvan_4639|M.vanbaalenii_PYR-1 RRD------YQPCCAAESYDAKQPQFEVPRRVVKGGSFLCADSYCQRYRP
. . : : :.:****.*** .**:***.
MAV_5033|M.avium_104 AARSSQSQDSATTHIGFRCIAS----
MSMEG_1226|M.smegmatis_MC2_155 AARSSQSQDSATTHIGFRCVAG----
MAB_4305|M.abscessus_ATCC_1997 AARSPQSQDSATTHIGFRCVENLG--
Mb0733|M.bovis_AF2122/97 AARSPQSQDTATTHIGFRCVADPVSG
Rv0712|M.tuberculosis_H37Rv AARSPQSQDTATTHIGFRCVADPVSG
Mflv_5038|M.gilvum_PYR-GCK SARSPQSQDSATTHIGFRCAASPV--
TH_0314|M.thermoresistible__bu SARSPQSQDSAATHIGFRCAA-----
MUL_0225|M.ulcerans_Agy99 AARRPQPIDTGVSHIGFRCVGRD---
MMAR_4408|M.marinum_M AARRPQPIDTGMSHIGFRCVGRD---
Mvan_4639|M.vanbaalenii_PYR-1 AARRPQPVDTGMSHIGFRCVKR----
:** .*. *:. :******