For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PIRNAAPLGAPCWIDLTTSDIDRAQDFYGTVFGWTFESAGPEYGGYVNAARNGHPVAGLMANNPEWQAPD NWTTYFHTADITAAAAAVTAAGGSRCVDPMEIPAKGFMCMATDPAGAFFGLWQPLGHRGFEIIGEAGAAV WHQLTTRDYHPAIGFYRRLFGWQTEQVGDTDEFRYTTASFDGEQLLGVMDGARLLREGVPSQWSIFFGAE DVDKTLQMIADNGGAVVRGAEDTPYGRLAAASDPTGANFNLSSLRA*
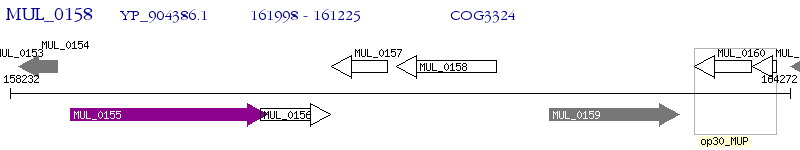
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0158 | - | - | 100% (257) | hypothetical protein MUL_0158 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0935 | - | 1e-119 | 74.32% (257) | hypothetical protein Mb0935 |
| M. gilvum PYR-GCK | Mflv_4018 | - | 6e-48 | 40.80% (250) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. tuberculosis H37Rv | Rv0911 | - | 1e-119 | 74.32% (257) | hypothetical protein Rv0911 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0976 | - | 2e-80 | 50.39% (258) | putative glyoxalase/bleomycin resistance protein |
| M. marinum M | MMAR_4617 | - | 1e-154 | 99.22% (257) | hypothetical protein MMAR_4617 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5616 | - | 1e-101 | 65.23% (256) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. thermoresistible (build 8) | TH_0683 | - | 2e-80 | 69.35% (186) | CONSERVED HYPOTHETICAL PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_4951 | - | 1e-107 | 68.87% (257) | glyoxalase/bleomycin resistance protein/dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0158|M.ulcerans_Agy99 ------------------------MPIRNAAPLGAPCWIDLTTSDIDRAQ
MMAR_4617|M.marinum_M ------------------------MPIRNAAPLGAPCWIDLTTSDIDRAQ
Mb0935|M.bovis_AF2122/97 ------------------------MPTRSSAPLGAPCWIDLTTSDVDRAQ
Rv0911|M.tuberculosis_H37Rv ------------------------MPTRSSAPLGAPCWIDLTTSDVDRAQ
MSMEG_5616|M.smegmatis_MC2_155 ------------------------MPIRSTAPLGAPSWIDLTTSDVERAK
Mvan_4951|M.vanbaalenii_PYR-1 MSFSRRGGLHGDRSPPLRRPEETAMPVATTAPLGAPIWIDLATSDLDRAQ
TH_0683|M.thermoresistible__bu ------------------------VPLRHGAPLGAPIWIDLGTSDLDRAL
MAB_0976|M.abscessus_ATCC_1997 ------------------------MPTRDETPLGAPAWIDLASSDLEKSK
Mflv_4018|M.gilvum_PYR-GCK -------------------------MSDYAAPVGAPIWLDLMSTDPARAA
:*:*** *:** ::* ::
MUL_0158|M.ulcerans_Agy99 DFYGTVFGWTFESA-GPEYGGYVNAARNGHPVAGLMANNPEWQAPDNWTT
MMAR_4617|M.marinum_M DFYGTVFGWTFESA-GPEYGGYVNAARNGHPVAGLMANNPEWQAPDNWTT
Mb0935|M.bovis_AF2122/97 DFYGTVFGWAFESA-GPDYGGYINAAKGGHPVAGLMANRPEFQSPDGWAT
Rv0911|M.tuberculosis_H37Rv DFYGTVFGWAFESA-GPDYGGYINAAKGGHPVAGLMANRPEFQSPDGWAT
MSMEG_5616|M.smegmatis_MC2_155 AFYGAVFGWTFETG-GPEYGGYVTAHLEGHVVAGLMQNDPQWNTPDVWTT
Mvan_4951|M.vanbaalenii_PYR-1 QFYGAVFGWTFESA-GPEYGGYVTAFRDDHPVAGLMVNDPQWNAPDAWTV
TH_0683|M.thermoresistible__bu QFYGAVFGWTFESA-GPDYGGYVNAFSQGRPVAGLMRNDPQWNAPDGWTT
MAB_0976|M.abscessus_ATCC_1997 AFYSALFGWTLQEA-GPEYGGYINAYKDGKAVAGMITNNPEWNAPDAWTT
Mflv_4018|M.gilvum_PYR-GCK EFYHAVFGWDVEAPPQAEFGGYQNFTVNGRRVAGLLPHMG--GVPNVWSV
** ::*** .: .::*** . .: ***:: : *: *:.
MUL_0158|M.ulcerans_Agy99 YFHTADITAAAAAVTAAGGSRCVDPMEIPAKGFMCMATDPAGAFFGLWQP
MMAR_4617|M.marinum_M YFHTADINAAAAAVTAAGGSRCVDPMEIPAKGFMCMATDPAGAFFGLWQP
Mb0935|M.bovis_AF2122/97 YFHTVDIGATVAKLAAAGGSSCLDPMEVPGKGFMSLAVDPSGAAFGLWQP
Rv0911|M.tuberculosis_H37Rv YFHTVDIGATVAKLAAAGGSSCLDPMEVPGKGFMSLAVDPSGAAFGLWQP
MSMEG_5616|M.smegmatis_MC2_155 YLHTADAQATVAAAAAAGGSNCGGVMDVPEKGRMAMMSDAAGGFFGLWQP
Mvan_4951|M.vanbaalenii_PYR-1 YLHTADIEATVAKATAAGGTSCVAPMEIPAKGWMGMLTDPTGAFLGLWQP
TH_0683|M.thermoresistible__bu YLHTADIHATLAAATAAGGTSCMAPMEVKDKGWMAMLSDPTGGVFGLWQP
MAB_0976|M.abscessus_ATCC_1997 YFATEDIDATLQKAVTHGGTNCLPPMEVPQLGYMALFADPAGAMVGLWQP
Mflv_4018|M.gilvum_PYR-GCK YLHTADAAETVRAVEAAGGSVMVPPMPVGDMGSMMVATDPAGAVIGFWQP
*: * * : : **: * : * * : *.:*. .*:***
MUL_0158|M.ulcerans_Agy99 LGHRGFEIIGEAGAAVWHQLTTRDYHPAIGFYRRLFGWQTEQV---GDTD
MMAR_4617|M.marinum_M LGHRGFEIIGEAGAAVWHQLTTRDYHPAIGFYRRLFGWQTEQV---GDTD
Mb0935|M.bovis_AF2122/97 LQHHGFEVIGEAGSPVWHQLTTRDYRSVIDFYRQVFGWRTEQI---SDTD
Rv0911|M.tuberculosis_H37Rv LQHHGFEVIGEAGSPVWHQLTTRDYRSVIDFYRQVFGWRTEQI---SDTD
MSMEG_5616|M.smegmatis_MC2_155 AGHDGFQVFNEPGAPLYHQLTTGDYAKALQFYRDVFGWTVNTV---ADTD
Mvan_4951|M.vanbaalenii_PYR-1 TGHRGYGIVGEHGAPVYHQLTTRGYGAALDFYRTVFGWTTETV---SDTD
TH_0683|M.thermoresistible__bu IGHQGFEVVGEPGAPVWHQLTTRDFGAALDFYRTVFGWQTQLV---SDTD
MAB_0976|M.abscessus_ATCC_1997 LAHKGFQLVGEGGAPVWHELNTREYDRAVDFYTTVLGWNTKIE---GDSA
Mflv_4018|M.gilvum_PYR-GCK GTHVGFTQWGVHGTPYWFECQSKDYEKSLAFYPQVIGARTEEIGTGGDPN
* *: . *:. :.: : : : ** ::* .: .*.
MUL_0158|M.ulcerans_Agy99 EF---RYTTASFDGEQLLGVMDGAR---LLREGVPSQWSIFFGAEDVDKT
MMAR_4617|M.marinum_M EF---RYTTASFDGEQLLGVMDGAR---LLPEGVPSQWSIFFGAEDVDKT
Mb0935|M.bovis_AF2122/97 EF---CYTTAWFDDQQLLGVMDGSS---CLPEGVPSNWTIFFGAEDVDET
Rv0911|M.tuberculosis_H37Rv EF---CYTTAWFDDQQLLGVMDGSS---CLPEGVPSNWTIFFGAEDVDET
MSMEG_5616|M.smegmatis_MC2_155 EF---RYSTASFDGDELVGVMDGAA---FLGD-APSTWTCFFGADDVDKT
Mvan_4951|M.vanbaalenii_PYR-1 EF---RYSTALFDGEALVGVMDGSN---IPGE-APSNWCFFLGADDVDKT
TH_0683|M.thermoresistible__bu EF---RYATAAFDGEDLX-XXGAAS---VAPCAAPAPSTP--RPVSAAPT
MAB_0976|M.abscessus_ATCC_1997 DS---RYACAQHDGAPIAGVNDASVGQWALPEGVPAHWAVYFGTDDTDAG
Mflv_4018|M.gilvum_PYR-GCK AVGPDRYAQLFIGESAYSGIMDSAT---LFPAEVPSFWQIYITVDDVAGT
*: . ..: .*: ..
MUL_0158|M.ulcerans_Agy99 LQMIADNGGAVVRGAEDTPYGRLAAASDPTGANFNLSSLRA--
MMAR_4617|M.marinum_M LQMIADNGGAVVRGAEDTPYGRLAAASDPTGANFNLSSLRA--
Mb0935|M.bovis_AF2122/97 LRVICDNGGSVVRAAENTPYGRLAAAADPMGVVFNLSSLQA--
Rv0911|M.tuberculosis_H37Rv LRVICDNGGSVVRAAENTPYGRLAAAADPMGVVFNLSSLQA--
MSMEG_5616|M.smegmatis_MC2_155 IELIVEHGGSVVRPAEDTPYGRLAAVADPTGAGFNLSSLQD--
Mvan_4951|M.vanbaalenii_PYR-1 VALVTDHGGSVVRGAEDTPYGRLAAVADPTGAGFNLSSIQS--
TH_0683|M.thermoresistible__bu VTTFHVAGRVMCLSFTRPSTPRLVHHRARR-------------
MAB_0976|M.abscessus_ATCC_1997 AAKAAELGGAVLAPAQDTPYGRMALIRDTTGGTFWLCSVDS--
Mflv_4018|M.gilvum_PYR-GCK VARAESLGAQILMPGEETPYGTLAALRDPLGALICLGHPPAGM
* : .. :.