For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGAGSGAMDQLSANRAASSEAAITGRHFKRLWGLPGTQLGVVSWPSERKDRMFGTWHYWWQAHLLDCLID AELRDPQAERRSRINRQVRSHRLRNNFSWTNSYYDDMAWLALALERAARIAGVQRRRALPKLADQFVKAW VPEDGGGIPWRKQDQFFNAPANGPAGIFLARYDDRLARAEQMGDWIDRTLIDPETHLVFDGIKGGSLVRA QYTYCQGVVLGLETELAARTGNDRHAPRVHRLVAAVAEHMASAGVLKGAGGGDGGLFAGITARYLALVAT TLPGDSADDAQARDTARAIVLASAKSAWDCRQTADGLPVFGAFWDRDAELPTAEGQQAQFVEGAVQGSEI AERDLSVQLSGWMLMEAAHSVAAVSSLG*
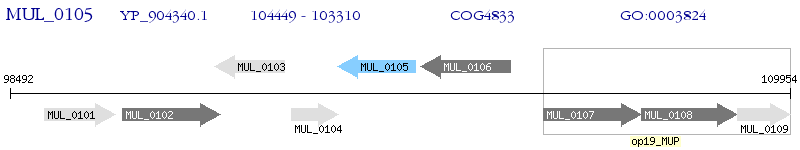
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0105 | - | - | 100% (379) | glycosyl hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0372c | - | 0.0 | 83.65% (373) | hypothetical protein Mb0372c |
| M. gilvum PYR-GCK | Mflv_0239 | - | 1e-156 | 72.07% (376) | glycoside hydrolase family protein |
| M. tuberculosis H37Rv | Rv0365c | - | 0.0 | 83.65% (373) | hypothetical protein Rv0365c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4257c | - | 1e-135 | 64.75% (366) | hypothetical protein MAB_4257c |
| M. marinum M | MMAR_0667 | - | 0.0 | 99.46% (371) | glycosyl hydrolase |
| M. avium 104 | MAV_4787 | - | 0.0 | 83.33% (372) | hypothetical protein MAV_4787 |
| M. smegmatis MC2 155 | MSMEG_0740 | - | 1e-172 | 77.51% (369) | glycosyl hydrolase family protein 76 |
| M. thermoresistible (build 8) | TH_0178 | - | 1e-159 | 73.46% (373) | CONSERVED HYPOTHETICAL PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_0666 | - | 1e-163 | 75.34% (369) | glycoside hydrolase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0239|M.gilvum_PYR-GCK --------MDQMWANRAASAEAAITTRHLRRVWGLPGTQLGVVAWPAATG
Mvan_0666|M.vanbaalenii_PYR-1 --------MDQMWANRAASAEAAISARHLRRLWGLPRTQLGVVAWPADGR
TH_0178|M.thermoresistible__bu --------MEQLWANRAASAEAAITQRHLRRLWGLPGTQLGVVAWPATGR
MUL_0105|M.ulcerans_Agy99 MSGAGSGAMDQLSANRAASSEAAITGRHFKRLWGLPGTQLGVVSWPSERK
MMAR_0667|M.marinum_M --------MDQLWANRAASSEAAITGRHFKRLWGLPGTQLGVVSWPSERK
Mb0372c|M.bovis_AF2122/97 --------MN--LANRAASAETAVTQRHLRRLWALPGTQLAVVAWPSTRR
Rv0365c|M.tuberculosis_H37Rv --------MN--LANRAASAETAVTQRHLRRLWALPGTQLAVVAWPSTRR
MAV_4787|M.avium_104 --------MDQLWANRAASCEAAVTQRHLRRLWGLPATQLGVVAWPPTRR
MSMEG_0740|M.smegmatis_MC2_155 --------MDQLWANRAASAEAAITERHLKKLWGLPGTQLGVVAWPPARK
MAB_4257c|M.abscessus_ATCC_199 --------MDVLWANRASSAEAAITARHLSPLWHLPGMQLGVVTWPPAPG
*: ****:*.*:*:: **: :* ** **.**:**.
Mflv_0239|M.gilvum_PYR-GCK HRRFGTWHYWWQAHLLDNLVDAQLRDPAPERLTRINRQIRGQRIRNIGKW
Mvan_0666|M.vanbaalenii_PYR-1 HRRFGTWHYWWQAHLLDNLVDAQVRDPRPERLSVITKQIRGLRLRNIGRW
TH_0178|M.thermoresistible__bu ERRFAIWHYWWQAHLLDCLVDAQLRDPQPDRLKRINRQIRGHRLRNMGRW
MUL_0105|M.ulcerans_Agy99 DRMFGTWHYWWQAHLLDCLIDAELRDPQAERRSRINRQVRSHRLRNNFSW
MMAR_0667|M.marinum_M DRMFGTWHYWWQAHLLDCLIDAELRDPQAERRSRINRQVRSHRLRNNFSW
Mb0372c|M.bovis_AF2122/97 DRLFGSWHYWWQAHLLDCLVDAQLRDPQPQRRARINRQVRSHRVRNNFSW
Rv0365c|M.tuberculosis_H37Rv DRLFGSWHYWWQAHLLDCLVDAQLRDPQPQRRARINRQVRSHRVRNNFSW
MAV_4787|M.avium_104 DRWFGTWHYWWQAHLLDCLVDAQQRDPQPQRRARINRQVRSHRLRNNLSW
MSMEG_0740|M.smegmatis_MC2_155 YTLFGTWHYWWQAHLLDCLVDAQVRDPQPERTTRIERQLRAHRLRNNLSW
MAB_4257c|M.abscessus_ATCC_199 ATWFKNWHYWWQAHLQDCLIDAQLRDPRPDRLKYIADMQRAHRFRNVFMW
* ********* * *:**: *** .:* * *. *.** *
Mflv_0239|M.gilvum_PYR-GCK TNSYYDDMAWLALALERAQRLTGVGRPKALDTLAEQFLTAWVPEDGGGIP
Mvan_0666|M.vanbaalenii_PYR-1 TNDYYDDMAWLALALERAGRLTGVTRPRALAKLADQFVTAWVPEDGGGIP
TH_0178|M.thermoresistible__bu TNDYYDDMAWLALALERSGRLTGVGRPKALDTLAGQLFSAWVPEDGGGIP
MUL_0105|M.ulcerans_Agy99 TNSYYDDMAWLALALERAARIAGVQRRRALPKLADQFVKAWVPEDGGGIP
MMAR_0667|M.marinum_M TNSYYDDMAWLALALERAARIAGVQRRRALPKLADQFVKAWVPEDGGGIP
Mb0372c|M.bovis_AF2122/97 LNSYYDDMAWLALALERADRVAGVRRRRALPKLTNQFVEAWVPEDGGGIP
Rv0365c|M.tuberculosis_H37Rv LNSYYDDMAWLALALERADRVAGVRRRRALPKLTNQFVEAWVPEDGGGIP
MAV_4787|M.avium_104 TNSYYDDMAWLALALERAARIAGVHRRKALPKLANQFLRAWVPEDGGGIP
MSMEG_0740|M.smegmatis_MC2_155 VNDYYDDMAWLALAIERAGRLAGVEKPTALRKLCDQFVTAWVPEDGGGIP
MAB_4257c|M.abscessus_ATCC_199 TNQYYDDMAWLALAMQRASELLGLERPRALNRLRQQFLDAWMPQYGGGIP
*.***********::*: .: *: : ** * *:. **:*: *****
Mflv_0239|M.gilvum_PYR-GCK WRKQDQFFNAPANGPAAIFLARHLAAQGESLRRAQQMADWIDETLLDPQT
Mvan_0666|M.vanbaalenii_PYR-1 WRKQDQFFNTPANGPAAIFLAR----YDDRLRRAQQMADWIDDTLIDPET
TH_0178|M.thermoresistible__bu WRRQDQFFNAPANGPAAIFLAR-----YDRLRRAEQMADWLHATLIDPET
MUL_0105|M.ulcerans_Agy99 WRKQDQFFNAPANGPAGIFLARYDD----RLARAEQMGDWIDRTLIDPET
MMAR_0667|M.marinum_M WRKQDQFFNAPANGPAGIFLARYDD----RLARAEQMGDWIDRTLIDPET
Mb0372c|M.bovis_AF2122/97 WRKQDQFFNAPANGPAGLFLARYPDQYGKRLKRAEQMADWIDRTLIDPET
Rv0365c|M.tuberculosis_H37Rv WRKQDQFFNAPANGPAGLFLARYPDQYGKRLKRAEQMADWIDRTLIDPET
MAV_4787|M.avium_104 WRKSDQFFNAPANGPAGIFLARYGD----HLKRAEQMANWIDETLIDPET
MSMEG_0740|M.smegmatis_MC2_155 WRKQDQFFNAPANGPAGIFLARYDD----RLRRAQQMADWIDETLIDPDT
MAB_4257c|M.abscessus_ATCC_199 WRKQDRFFNAPANGPAAIVLARTG-----RVWRAEVMSDWIDKTLVDPDS
**:.*:***:******.:.*** : **: *.:*:. **:**::
Mflv_0239|M.gilvum_PYR-GCK HLIFDGIKG-GSLVRAQYTYCQGVVLGVETELAARTS---DSRHAERVHR
Mvan_0666|M.vanbaalenii_PYR-1 HLVFDGIKA-GSLVRAQYTYCQGVVLGLETELAVRTE---DARHPERVHR
TH_0178|M.thermoresistible__bu HLVFDGIKA-GSLVRAQYTYCQGVVLGLEAELAVRTG---NPVHARRVYR
MUL_0105|M.ulcerans_Agy99 HLVFDGIKG-GSLVRAQYTYCQGVVLGLETELAARTG---NDRHAPRVHR
MMAR_0667|M.marinum_M HLVFDGIKG-GSLVRAQYTYCQGVVLGLETELAARTG---NDRHAPRVHR
Mb0372c|M.bovis_AF2122/97 HLVFDGIKA-GSLVRAQYTYCQGVVLGLETELAVRTGPAARARHCARVHR
Rv0365c|M.tuberculosis_H37Rv HLVFDGIKA-GSLVRAQYTYCQGVVLGLETELAVRTGPAARARHCARVHR
MAV_4787|M.avium_104 HLVFDGIKA-GSLVRAQYTYCQGVVLGLETELAARTDGPARQRHAARVRR
MSMEG_0740|M.smegmatis_MC2_155 HLVFDGIKA-GSLVRAQYTYCQGVVLGLEVELAVRTQ---DPRHAPRVRR
MAB_4257c|M.abscessus_ATCC_199 HLVIDGIQEDGTLERATYTYCQGVVLGLETELAVRTA---EPRHAQRVHR
**::***: *:* ** **********:*.***.** * ** *
Mflv_0239|M.gilvum_PYR-GCK LVAAVAEHMAPGGVINGAGGGDGGLFNGILARYLALVATALPGTSPADEQ
Mvan_0666|M.vanbaalenii_PYR-1 LVAAVAEHMAPAGVIKGAGGGDGGLFNGILARYLALVATALPGGSPADDK
TH_0178|M.thermoresistible__bu LVAAVRDHMAPDGVIKGAGGGDGGLFNAILARYLALVAQILPDGPGADAE
MUL_0105|M.ulcerans_Agy99 LVAAVAEHMASAGVLKGAGGGDGGLFAGITARYLALVATTLPGDSADDAQ
MMAR_0667|M.marinum_M LVAAVAEHMASAGVLKGAGGGDGGLFAGITARYLALVTTTLPGDSADDAQ
Mb0372c|M.bovis_AF2122/97 LVAAVNEHMAPLGVLRGAGGGDGGLFAGITARYLALVATTLPGDSADDAA
Rv0365c|M.tuberculosis_H37Rv LVAAVNEHMAPLGVLRGAGGGDGGLFAGITARYLALVATTLPGDSADDAA
MAV_4787|M.avium_104 LVAAVAEHMAPAGVLAGAGGGDGGLFAGVTARYLALVATTLPGSAAEDVA
MSMEG_0740|M.smegmatis_MC2_155 LVAAVDKEMTTDGVIKGAGGGDGGLFNGILARYLALVATTLPGETPEDIA
MAB_4257c|M.abscessus_ATCC_199 LVRAVRDLMTEDGVIRGSGGGDGGLFNGILARYLALVVTLLPQNSPQDAD
** ** . *: **: *:******** .: *******. ** . *
Mflv_0239|M.gilvum_PYR-GCK A----RRTATELVLASGRAAWDNRQTGKDTEDLPLFGAFWDRPAQVPTAG
Mvan_0666|M.vanbaalenii_PYR-1 A----RETATELVLTSARTAWDNRQT---VEGLPLFAAFWDRPAELPTAG
TH_0178|M.thermoresistible__bu ASRTTRETARTLVLDSARSAWDHRQV---VDGLPLFGPFWDRAAELPRAG
MUL_0105|M.ulcerans_Agy99 A----RDTARAIVLASAKSAWDCRQT---ADGLPVFGAFWDRDAELPTAE
MMAR_0667|M.marinum_M A----RDTARAIVLASAKSAWDCRQT---ADGLPVFGAFWDRDAELPTAE
Mb0372c|M.bovis_AF2122/97 A----RDTARAIVLASAQSAWDYRQT---VDGLPVFGAFWDREAELPTAG
Rv0365c|M.tuberculosis_H37Rv A----RDTARAIVLASAQSAWDYRQT---VDGLPVFGAFWDREAELPTAG
MAV_4787|M.avium_104 A----RDTARRIVLASAKSAWDYRQT---LDGLPVFGPFWDRDAQLPTAG
MSMEG_0740|M.smegmatis_MC2_155 A----RDTARTIVLKSARSAWDHRQT---VDGLPLFSAFWDRNAEIPTAG
MAB_4257c|M.abscessus_ATCC_199 A----RATARDILLASAEAAWANRLT---VESLPLFGADWTRTADLPTAQ
* * ** ::* *..:** * . :.**:*.. * * *::* *
Mflv_0239|M.gilvum_PYR-GCK TADARSVDGAVNSSEIPERDLSVQLAGWMLMEAACVVTAGDSD-------
Mvan_0666|M.vanbaalenii_PYR-1 SGDAELVDGAVNSSEIPERDLSVQLAGWMLLEAAHVVTAGDSD-------
TH_0178|M.thermoresistible__bu GREAQFVEGAVNASEIPERDLSVQLSGWMLMEAAHALTDETSAETSTDET
MUL_0105|M.ulcerans_Agy99 GQQAQFVEGAVQGSEIAERDLSVQLSGWMLMEAAHSVAAVSSLG------
MMAR_0667|M.marinum_M GQQAQFVEGAVQGSEIAERDLSVQLSGWMLMEAAHSVAAVSSLG------
Mb0372c|M.bovis_AF2122/97 GEQARSVRGAVHSSAIAERDLSVQLSGWMLMEAAHSAAAVSSLG------
Rv0365c|M.tuberculosis_H37Rv GEQARSVRGAVHSSAIAERDLSVQLSGWMLMEAAHSAAAVSSLG------
MAV_4787|M.avium_104 GKQAEFVEGAVTASEIAERDLSVQLSGWMLMEAACNVSAESSHENRSAL-
MSMEG_0740|M.smegmatis_MC2_155 GKEAEFVEGAVNASSVPERDLSVQLAGWMLMEAAHSVDSVDAEDAAGDQN
MAB_4257c|M.abscessus_ATCC_199 SAASQFVAGAVNPSKVPERDLSVQLSGWMLLEAAHVVA------------
:. * *** * :.********:****:***
Mflv_0239|M.gilvum_PYR-GCK ----------------
Mvan_0666|M.vanbaalenii_PYR-1 ----------------
TH_0178|M.thermoresistible__bu ADEADEEENPA-----
MUL_0105|M.ulcerans_Agy99 ----------------
MMAR_0667|M.marinum_M ----------------
Mb0372c|M.bovis_AF2122/97 ----------------
Rv0365c|M.tuberculosis_H37Rv ----------------
MAV_4787|M.avium_104 ----------------
MSMEG_0740|M.smegmatis_MC2_155 GAGDGEVGHDENEDEE
MAB_4257c|M.abscessus_ATCC_199 ----------------