For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KHGSVPATPEAASLVFGDRLEAAELYARILAGAGVEWGLLGPREVDRVWERHILNSAALGELMEPGERVA DIGSGAGLPGIPLALARPDIHVTLIEPLLRRSEFLRETVTELGLDVTVVRGRAEDREVRDRVGEMDVVTS RAVASLDKLTRWSVPFLRDGGRMLPIKGERAEVEIEEHRRVMESLGAVDARVVRCGANYLSPPVTVVDAR RRAAKPGRNKSGRTARSRGRTGRR*
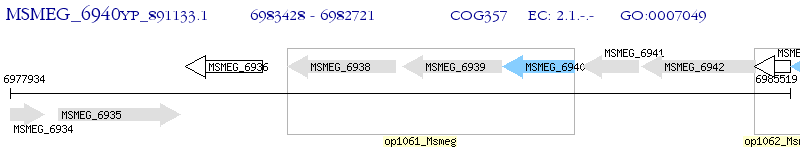
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6940 | gidB | - | 100% (235) | methyltransferase GidB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3950c | gidB | 2e-74 | 64.22% (218) | 16S rRNA methyltransferase GidB |
| M. gilvum PYR-GCK | Mflv_0833 | gidB | 7e-83 | 66.08% (227) | methyltransferase GidB |
| M. tuberculosis H37Rv | Rv3919c | gidB | 2e-73 | 63.76% (218) | 16S rRNA methyltransferase GidB |
| M. leprae Br4923 | MLBr_02708 | gidB | 2e-73 | 61.93% (218) | 16S rRNA methyltransferase GidB |
| M. abscessus ATCC 19977 | MAB_4951c | gidB | 3e-71 | 58.37% (233) | methyltransferase GidB (glucose-inhibited division protein |
| M. marinum M | MMAR_5483 | gidB | 7e-79 | 68.37% (215) | glucose-inhibited division protein B Gid |
| M. avium 104 | MAV_5308 | gidB | 3e-78 | 65.18% (224) | glucose-inhibited division protein B |
| M. thermoresistible (build 8) | TH_0763 | gid | 3e-87 | 69.57% (230) | PROBABLE GLUCOSE-INHIBITED DIVISION PROTEIN B GID |
| M. ulcerans Agy99 | MUL_5072 | gid | 3e-56 | 66.46% (158) | glucose-inhibited division protein B Gid |
| M. vanbaalenii PYR-1 | Mvan_6073 | gidB | 4e-87 | 69.16% (227) | methyltransferase GidB |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0833|M.gilvum_PYR-GCK -MKHAEIPPPPDAAA------------------EVFGPALGAARRYAEIL
Mvan_6073|M.vanbaalenii_PYR-1 -MKHAEVPPPPEAAE------------------AVFGPALEKAHLYARIL
MSMEG_6940|M.smegmatis_MC2_155 -MKHGSVPATPEAAS------------------LVFGDRLEAAELYARIL
TH_0763|M.thermoresistible__bu -VKHGAVAAPPPAAA------------------TIFGPALDRAQRYAAIL
MMAR_5483|M.marinum_M -----MTQPLDDAAS------------------AIFGPRLELAQRYADWL
MUL_5072|M.ulcerans_Agy99 -----MTQPLDDAAS------------------AIFGPRLELAQCYADWL
Mb3950c|M.bovis_AF2122/97 ------MSPIEPAAS------------------AIFGPRLGLARRYAEAL
Rv3919c|M.tuberculosis_H37Rv ------MSPIEPAAS------------------AIFGPRLGLARRYAEAL
MAV_5308|M.avium_104 ---MKHVGPVEPAAGGPEVPPVAALGAAPESAAALFGPRLATAQRYAEVL
MLBr_02708|M.leprae_Br4923 MFHVKHVGSYEELASFTSVKGNLGFDTVFEAVFMIFGPRLNIAQRYVDLL
MAB_4951c|M.abscessus_ATCC_199 -MKHGETLPVPEAAA------------------EVFGERLARAVEYVAAL
. * :** * * *. *
Mflv_0833|M.gilvum_PYR-GCK AGAGVERGLLGPREVDRIWDRHILNCAVIGELVEAGERVADVGSGAGLPG
Mvan_6073|M.vanbaalenii_PYR-1 AGAGVERGLLGPREVERLWDRHILNCAVVGELLQPGERVADIGSGAGLPG
MSMEG_6940|M.smegmatis_MC2_155 AGAGVEWGLLGPREVDRVWERHILNSAALGELMEPGERVADIGSGAGLPG
TH_0763|M.thermoresistible__bu AGAGVERGLLGPREVDRIWDRHVLNSAVTSELIEPGERVADIGSGAGLPG
MMAR_5483|M.marinum_M ATAGVERGLLGPREVDRLWERHVLNSAVIGELLDQGERVVDIGSGAGLPG
MUL_5072|M.ulcerans_Agy99 GTAGVERGLLGPREVDRLWERHVLNSAVIGELLDHGERVVDIGSGAGLPA
Mb3950c|M.bovis_AF2122/97 AGPGVERGLVGPREVGRLWDRHLLNCAVIGELLERGDRVVDIGSGAGLPG
Rv3919c|M.tuberculosis_H37Rv AGPGVERGLVGPREVGRLWDRHLLNCAVIGELLERGDRVVDIGSGAGLPG
MAV_5308|M.avium_104 GTAGVERGLLGPREVDRIWDRHILNSAAVAGLLGRGDRIIDIGSGAGLPG
MLBr_02708|M.leprae_Br4923 ANTGIERGLLGPHEANRLWDRHLLNSAVVAELLDPGDRVVDIGSGAGLPG
MAB_4951c|M.abscessus_ATCC_199 ATIGVERGLIGPRETGRLWGRHILNSAVLGELLEAGERVIDVGSGAGLPG
. *:* **:**:*. *:* **:**.*. . *: *:*: *:*******.
Mflv_0833|M.gilvum_PYR-GCK IPLALARPDVHVVLIEPLLRRSDFLREAIEDVGIE-CSVVRGRAEDRSVR
Mvan_6073|M.vanbaalenii_PYR-1 IPLALARPDVHVILIEPLLRRSEFLRETIEEIGID-CDVVRGRAEDRSVR
MSMEG_6940|M.smegmatis_MC2_155 IPLALARPDIHVTLIEPLLRRSEFLRETVTELGLD-VTVVRGRAEDREVR
TH_0763|M.thermoresistible__bu IPLALARPDLSITLVEPMLRRSEFLHEVVAELDLE-VRVVRGRAEDPAVR
MMAR_5483|M.marinum_M LPLAIARPDLQVVLLEPMLRRVEFLQEVVTDLGLA-VEVVRGRAEERSVR
MUL_5072|M.ulcerans_Agy99 LPLAIARHDLQVVLLEPMLRRVEFLQEVVTDLGLA-VEVVRGRAEERSVR
Mb3950c|M.bovis_AF2122/97 VPLAIARPDLQVVLLEPLLRRTEFLREMVTDLGVA-VEIVRGRAEESWVQ
Rv3919c|M.tuberculosis_H37Rv VPLAIARPDLQVVLLEPLLRRTESLREMVTDLGVA-VEIVRGRAEESWVQ
MAV_5308|M.avium_104 IPLAIARPDLEVVLLEPLLRRSEFLTEVVDELGLA-VEVVRGRAEERPVR
MLBr_02708|M.leprae_Br4923 LPLAIARPDLQVVLLEPLLRRVTFLREVVAELGLD-VEVVRGRAEELWVR
MAB_4951c|M.abscessus_ATCC_199 IPLGIARPDVGIVLVEPMLRRTEFLREMIEQLGLTNVSVVRGRAEEPAVI
:**.:** *: : *:**:*** * * : ::.: :******: *
Mflv_0833|M.gilvum_PYR-GCK EEVGPTDVVVSRAVASLDKLAKWSSPLLRPGGRMLAIKGERAAEEIEEHR
Mvan_6073|M.vanbaalenii_PYR-1 EEVGTTDVVVSRAVASLDKLTKWSSPLLRVGGRMLAIKGERADDEVQQHR
MSMEG_6940|M.smegmatis_MC2_155 DRVGEMDVVTSRAVASLDKLTRWSVPFLRDGGRMLPIKGERAEVEIEEHR
TH_0763|M.thermoresistible__bu REVGEMDAVTSRAVAPLDKLARWSLPLLKPGGHMLAIKGARAEAEVDEHR
MMAR_5483|M.marinum_M ERLGGSDAAVSRAVAALDKLTKWSMPLLKREGRMLAIKGERAPDEVREHR
MUL_5072|M.ulcerans_Agy99 ERLGGSDAAVSRAVAALDKLAKWSMPLLKREGRMLAIK------------
Mb3950c|M.bovis_AF2122/97 DQLGGSDAAVSRAVAALDKLTKWSMPLIRPNGRMLAIKGERAHDEVREHR
Rv3919c|M.tuberculosis_H37Rv DQLGGSDAAVSRAVAALDKLTKWSMPLIRPNGRMLAIKGERAHDEVREHR
MAV_5308|M.avium_104 NRFGDRDAAVSRAVAALDKLTKWSMPLLRHDGRMLAIKGERAAEEVDRYR
MLBr_02708|M.leprae_Br4923 DRIGERDVAVSRAVAALDKLTKWSIPLLRPGGQILAIKGEHVFDEIHQHR
MAB_4951c|M.abscessus_ATCC_199 AEVGGADAVTSRAVAALDKIARWSLPLTRIGGRMLAIKGDRAEDEVEQYG
..* *...*****.***:::** *: : *::*.**
Mflv_0833|M.gilvum_PYR-GCK RALAALGVSELKVERCGARFVDPPATVVVGFQATAPEK----RPRSGRRQ
Mvan_6073|M.vanbaalenii_PYR-1 RAMAALGVSEVKVERCGGQYVEPPATVVVGFQGVAKVS----QSRSGRRH
MSMEG_6940|M.smegmatis_MC2_155 RVMESLGAVDARVVRCGANYLSPPVTVVDARRRAAKPG----RNKSGRTA
TH_0763|M.thermoresistible__bu RVLNRLKAVDVRVVRCGADLLDPPATVVVARRAAAKQGGYP-PTRSGRRR
MMAR_5483|M.marinum_M RVMESLGAADVRVVPCGANYLQPPATVVVARRGDTRGPNRR-VSPRRTGG
MUL_5072|M.ulcerans_Agy99 -----------------------------------RGASSR---------
Mb3950c|M.bovis_AF2122/97 RVMIASGAVDVRVVTCGANYLRPPATVVFARRG--KQIARG-SARMASGG
Rv3919c|M.tuberculosis_H37Rv RVMIASGAVDVRVVTCGANYLRPPATVVFARRG--KQIARG-SARMASGG
MAV_5308|M.avium_104 RVMTASGAADVRVVTCGANYLRPPATVVSARRA--KPPHPK-SARTGKAG
MLBr_02708|M.leprae_Br4923 RVMASLGAVDVMVVVCGANYLCRPVTVVLTRCG--QQMRHK-PARVGDRK
MAB_4951c|M.abscessus_ATCC_199 SALGQLGADDVRVVQCGTRYLDSPTTVVVARRKECDGNRQRGSSRGGSRK
Mflv_0833|M.gilvum_PYR-GCK R--------
Mvan_6073|M.vanbaalenii_PYR-1 R--------
MSMEG_6940|M.smegmatis_MC2_155 RSRGRTGRR
TH_0763|M.thermoresistible__bu G--------
MMAR_5483|M.marinum_M APA------
MUL_5072|M.ulcerans_Agy99 ---------
Mb3950c|M.bovis_AF2122/97 TA-------
Rv3919c|M.tuberculosis_H37Rv TA-------
MAV_5308|M.avium_104 TR-------
MLBr_02708|M.leprae_Br4923 TQ-------
MAB_4951c|M.abscessus_ATCC_199 RSR------