For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VIPVLELAILGLLLESPMHGYELRKRLTGLLGAFRAFSYGSLYPALRRMQADGLIVEDAAPEGATKVRRA RRVYQLTEAGKQRFAELVADTGPQNYSDDGFGVHLAFFNRTPAEARMRILEGRRRQVEERREGLREAVAR ASSSFDRYTRQLHQLGLESSEREVKWLNELIAAERLSQGRTDNP
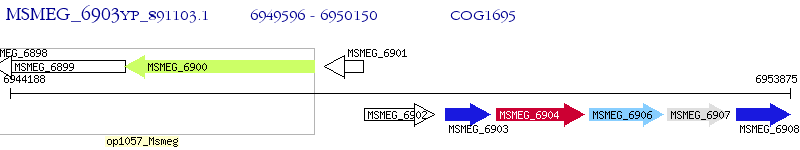
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6903 | - | - | 100% (184) | transcriptional regulator, PadR family protein |
| M. smegmatis MC2 155 | MSMEG_6227 | - | 2e-06 | 36.49% (74) | transcriptional regulator, PadR family protein |
| M. smegmatis MC2 155 | MSMEG_6226 | - | 3e-06 | 25.86% (174) | transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0048c | - | 5e-86 | 91.86% (172) | hypothetical protein Mb0048c |
| M. gilvum PYR-GCK | Mflv_0868 | - | 2e-90 | 91.62% (179) | PadR-like family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0047c | - | 5e-86 | 91.86% (172) | hypothetical protein Rv0047c |
| M. leprae Br4923 | MLBr_02691 | - | 9e-83 | 84.92% (179) | hypothetical protein MLBr_02691 |
| M. abscessus ATCC 19977 | MAB_4906 | - | 6e-86 | 86.26% (182) | PadR family transcriptional regulator |
| M. marinum M | MMAR_0066 | - | 5e-85 | 87.15% (179) | hypothetical protein MMAR_0066 |
| M. avium 104 | MAV_0068 | - | 2e-86 | 88.83% (179) | transcriptional regulator, PadR family protein |
| M. thermoresistible (build 8) | TH_1310 | - | 3e-93 | 90.76% (184) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0065 | - | 3e-85 | 87.15% (179) | hypothetical protein MUL_0065 |
| M. vanbaalenii PYR-1 | Mvan_6039 | - | 1e-92 | 93.85% (179) | PadR-like family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0048c|M.bovis_AF2122/97 ---MLELAILGLLIESPMHGYELRKRLTGLLGAFRAFSYGSLYPALRRMQ
Rv0047c|M.tuberculosis_H37Rv ---MLELAILGLLIESPMHGYELRKRLTGLLGAFRAFSYGSLYPALRRMQ
MMAR_0066|M.marinum_M ---MLELAILGLLIESPMHGYELRKRLTGLLGAFRAFSYGSLYPALRRMQ
MUL_0065|M.ulcerans_Agy99 ---MLELAILGLLIESPMHGYELRKRLTGLLGAFRAFSYGSLYPALRRMQ
MAV_0068|M.avium_104 ---MLELAILGLLIESPMHGYELRKRLTGLLGAFRAFSYGSLYPALRRMQ
MLBr_02691|M.leprae_Br4923 ---MLELAILGLLIESPMHGYELRKRLTGLLGAFRAFSYGSLYPALRRMQ
Mflv_0868|M.gilvum_PYR-GCK ---MLELAILGLLQESPMHGYELRKRLTGLLGAFRAFSYGSLYPALRRMQ
Mvan_6039|M.vanbaalenii_PYR-1 ---MLELAILGLLQESPMHGYELRKRLTGLLGAFRAFSYGSLYPALRRMQ
TH_1310|M.thermoresistible__bu VIRLLELAILGLLLESPMHGYELRKRLTGLLGAFRAFSYGSLYPALRRMQ
MSMEG_6903|M.smegmatis_MC2_155 MIPVLELAILGLLLESPMHGYELRKRLTGLLGAFRAFSYGSLYPALRRMQ
MAB_4906|M.abscessus_ATCC_1997 ---MLELAILGLLLESPMHGYELRKRLTGLLGAFRAFSYGSLYPALRRMQ
:********* ************************************
Mb0048c|M.bovis_AF2122/97 ADGLIAENAAP---AGTP-VRRARRVYQLTDKGRRRFGELVADTGPHNYT
Rv0047c|M.tuberculosis_H37Rv ADGLIAENAAP---AGTP-VRRARRVYQLTDKGRRRFGELVADTGPHNYT
MMAR_0066|M.marinum_M AEGLIAENAAP---TGMP-VRRARRVYQLTDAGRRRFGELVADTGPHNYT
MUL_0065|M.ulcerans_Agy99 AEGLIAENAAP---TGMP-VRRARRVYQLTDAGRRRFGELVADTGPHNYT
MAV_0068|M.avium_104 AEGLIAEDAAP---AGTP-VRRARRVYQLTDEGRRRFGELVADTGPHNYT
MLBr_02691|M.leprae_Br4923 ADRLITGHVAP---AGTP-VRRARQVYQLTDAGRRRFSELVTDTGPDNYT
Mflv_0868|M.gilvum_PYR-GCK TDGLIVEDSAP---EGTVKVRRARRVYQLTESGMQRFTELVADTGPQNYS
Mvan_6039|M.vanbaalenii_PYR-1 ADGLIVEDAAP---EGTVKVRRARRVYQLTDAGKQRFTELVADTGPQNYS
TH_1310|M.thermoresistible__bu ADGLIVEDPAP---DGTPKVRRARRVYRLTEAGKQRFTELVADTGPQNYS
MSMEG_6903|M.smegmatis_MC2_155 ADGLIVEDAAP---EGATKVRRARRVYQLTEAGKQRFAELVADTGPQNYS
MAB_4906|M.abscessus_ATCC_1997 ADGLIEEDAAPGINSAATMLRRGKRVYQLTTAGRSRFSELVADTGPQNYT
:: ** . ** . :**.::**:** * ** ***:****.**:
Mb0048c|M.bovis_AF2122/97 DDGFGVHLAFFNRTPAEARMRILEGRRRQVEERREGLREAVARASSSFDR
Rv0047c|M.tuberculosis_H37Rv DDGFGVHLAFFNRTPAEARMRILEGRRRQVEERREGLREAVARASSSFDR
MMAR_0066|M.marinum_M DDGFGVHLAFFNRTPAEARMRILEGRRRQVEERREGLRDAVARASSSFDR
MUL_0065|M.ulcerans_Agy99 DDGFGVHLAFFNRTPAEARMRILEGRRRQVEERREGLRDAVARASSSFDR
MAV_0068|M.avium_104 DDGFGVHLAFFNRTPAEARMRILEGRRRQVEERREGLREAVARASSSFDR
MLBr_02691|M.leprae_Br4923 DNGFGVHLAFFNRTPAEARMRILEGRRRRVEERREGLREAVTRASNSFDR
Mflv_0868|M.gilvum_PYR-GCK DDGFGVHLAFFNRTPAEARMRILEGRRRQVEERREGLREAVARASNSFDR
Mvan_6039|M.vanbaalenii_PYR-1 DDGFGVHLAFFNRTPAEARMRILEGRRRQVEERREGLRDAVARASSSFDR
TH_1310|M.thermoresistible__bu DDGFGVHLAFFNRTPAEARMRILEGRRRQVEERREGLREAIARASNSFDR
MSMEG_6903|M.smegmatis_MC2_155 DDGFGVHLAFFNRTPAEARMRILEGRRRQVEERREGLREAVARASSSFDR
MAB_4906|M.abscessus_ATCC_1997 DDGFGVHLAFFNRTPAEARMRILEGRRRQVEERREGLREAITRASSSFDR
*:**************************:*********:*::***.****
Mb0048c|M.bovis_AF2122/97 YTRQLHQLGLESSEREVKWLNELIAAERAAPNPAEQT
Rv0047c|M.tuberculosis_H37Rv YTRQLHQLGLESSEREVKWLNELIAAERAAPNPAEQT
MMAR_0066|M.marinum_M YTRQLHQLGLESSEREVKWLNELIAAERAAPNHAEQT
MUL_0065|M.ulcerans_Agy99 YTRQLHQLGLESSEREVKWLNELIAAERAAPNHAEQT
MAV_0068|M.avium_104 YTRQLHQLGLESSEREVKWLNELIAAERAMPGLSEQT
MLBr_02691|M.leprae_Br4923 YTRQLHQLGLESSEREVKWLNELIAAERALLGRVEQT
Mflv_0868|M.gilvum_PYR-GCK YTRQLHQLGLESSEREVTWLNELIAAEKSAQGRAEQT
Mvan_6039|M.vanbaalenii_PYR-1 YTRQLHQLGLESSEREVKWLNELIAAERSAQGRAEQT
TH_1310|M.thermoresistible__bu YTRQLHQLGLESSEREVNWLNDLIAAERMAQGRAEQP
MSMEG_6903|M.smegmatis_MC2_155 YTRQLHQLGLESSEREVKWLNELIAAERLSQGRTDNP
MAB_4906|M.abscessus_ATCC_1997 YTKQLHQLGLESSEREVKWLNDLIAAERTAQARVEER
**:**************.***:*****: ::