For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVRQLASLTRGAVRTLATDGAVAVLPIGAVEQHGDHLPLGTDFLLVEAVCDRALQQLEDDRTTWVRVPT MTFGHSPHHLFAAAVSLQAGTLTDVLADVLDSLAQTGFRRIVIVNGHGGNDELVRLAVKAHSLHAEGTAA ACSYWTLTAQTTAEAAATERPDRTPGHAGWFETSLMLAVHPELVADDRPFTPSDPPPLFNRSPAPGLTVE RHGEWPRVGGTTDDAREASAETGRLLLGRRVAGLVAALQSLDRATATPPAASVPSTDRERGPQA
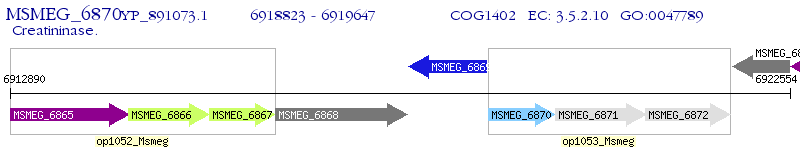
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6870 | - | - | 100% (274) | creatinine amidohydrolase |
| M. smegmatis MC2 155 | MSMEG_1425 | - | 1e-13 | 34.13% (167) | creatininase subfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0714 | - | 4e-15 | 32.75% (229) | hypothetical protein Mb0714 |
| M. gilvum PYR-GCK | Mflv_5058 | - | 2e-18 | 32.30% (226) | creatininase |
| M. tuberculosis H37Rv | Rv0695 | - | 4e-15 | 32.75% (229) | hypothetical protein Rv0695 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3832c | - | 8e-16 | 36.99% (173) | hypothetical protein MAB_3832c |
| M. marinum M | MMAR_1023 | - | 3e-17 | 32.03% (231) | creatinine amidohydrolase |
| M. avium 104 | MAV_4476 | - | 5e-16 | 33.03% (221) | creatininase subfamily protein |
| M. thermoresistible (build 8) | TH_1902 | - | 4e-07 | 30.54% (167) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0775 | - | 8e-18 | 32.47% (231) | creatinine amidohydrolase |
| M. vanbaalenii PYR-1 | Mvan_1602 | - | 2e-19 | 35.79% (190) | creatininase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0714|M.bovis_AF2122/97 MNSSYHRRVPVVGELGSATSSQLPS----TSPSIVIPLGSTEQHGPHLPL
Rv0695|M.tuberculosis_H37Rv MNSSYHRRVPVVGELGSATSSQLPS----TSPSIVIPLGSTEQHGPHLPL
MMAR_1023|M.marinum_M MNSSYHRRVPVLGELGTSTSSQLPS----TSPSILIPLGSTEQHGPHLPL
MUL_0775|M.ulcerans_Agy99 MNSSYHRRVPVLGELGTSTSSQLPS----TWPSILIPLGSTEQHGPHLPL
MAV_4476|M.avium_104 MNSSYHHRVSVLGELGTVTSSQLSG----TAVSLLVPLGSTEQHGPHLPL
Mflv_5058|M.gilvum_PYR-GCK MNSAYHRRVVFPRELGNSTSRQLHDAHAIHMPTLVVPVGSTEQHGPHLPL
TH_1902|M.thermoresistible__bu -------------------------------------------------L
MAB_3832c|M.abscessus_ATCC_199 MNSAYHRRVAVPTVLSTAISPELSD----EAITLFTPLGSTEQHGPHLPL
Mvan_1602|M.vanbaalenii_PYR-1 --------MPNWGDLSRDEVADAAR----SGAVAVLPVGAIEQHGAHLPT
MSMEG_6870|M.smegmatis_MC2_155 ------MTVRQLASLTRGAVRTLAT----DGAVAVLPIGAVEQHGDHLPL
Mb0714|M.bovis_AF2122/97 DTDTRIATAVARTVTARLHAEDLPIAQEEWLMAPAIAYGASGEHQR-FAG
Rv0695|M.tuberculosis_H37Rv DTDTRIATAVARTVTARLHAEDLPIAQEEWLMAPAIAYGASGEHQR-FAG
MMAR_1023|M.marinum_M DTDTRIATAVGRAVATRMHGR-LTQCQPGWLLAPPIAYGASGEHQS-FAG
MUL_0775|M.ulcerans_Agy99 DTDTRIATAVGRAVATRMHGR-LTQCQPGWLLAPPIAYGASGEHQS-FAG
MAV_4476|M.avium_104 DTDTRIASAVAGRAAPRLGDG--------WLVAPAIAYGASGEHQS-FAG
Mflv_5058|M.gilvum_PYR-GCK DTDTRIATAVADAVVTRLRARD----ETHWVMAPAIAYGASGEHEG-FPG
TH_1902|M.thermoresistible__bu DTDTRIAGAVARAVIERLTAADG---DTRWLLAPPICYGASGEHEG-FPG
MAB_3832c|M.abscessus_ATCC_199 DTDTRIAGAVATGAAAELAADGG---DGRTALAPAIAYGASGEHQS-FAG
Mvan_1602|M.vanbaalenii_PYR-1 GTDTLLATAAVSAATARTGDVG----------LPAVSYGCSLGHTAHWPG
MSMEG_6870|M.smegmatis_MC2_155 GTDFLLVEAVCDRALQQLEDD-----RTTWVRVPTMTFGHS--PHHLFAA
.** :. *. . . *.: :* * :..
Mb0714|M.bovis_AF2122/97 TISIGTEALTMLLVEYGRSAAC-WARRLVFVNGHGGNVGALTRAVGLLRA
Rv0695|M.tuberculosis_H37Rv TISIGTEALTMLLVEYGRSAAC-WARRLVFVNGHGGNVGALTRAVGLLRA
MMAR_1023|M.marinum_M TISIGTEALRVLLLEYGRSAAC-WADRLVFVNGHGGNVEALRGAVRQLRA
MUL_0775|M.ulcerans_Agy99 TISIGAEALRVLLLEYGRSAAC-WADRLVFVNGHGGNVEALRGAVRQLRA
MAV_4476|M.avium_104 TVSIGTEALTLLLIEYGRSAAG-WAQRVVFVNGHGGNVAALGRAVGTLRA
Mflv_5058|M.gilvum_PYR-GCK TISIGTPALAELLVEFVRSACR-WTPRVAFVNGHGGNVDALRRAVTQLRH
TH_1902|M.thermoresistible__bu TVSVGTPALRLWLVEYGRSAAN-WASRLVFLNGHGGNVEALAAATALLRY
MAB_3832c|M.abscessus_ATCC_199 TISIGTEVLGRLLVEYARSAIQ-WCRRIVIVNGHGGNIQGLVSAVRLLRS
Mvan_1602|M.vanbaalenii_PYR-1 TLSLSVSTMTALVTDIGRWVYTSGFRKLVIVNAHATNGPPTQGALLTLRH
MSMEG_6870|M.smegmatis_MC2_155 AVSLQAGTLTDVLADVLDSLAQTGFRRIVIVNGHGGNDELVRLAVKAHSL
::*: . .: : : ::.::*.*. * *
Mb0714|M.bovis_AF2122/97 EG-------------RDAGWCPCTCPGGD---PHAGHTETSVLLHLSPAD
Rv0695|M.tuberculosis_H37Rv EG-------------RDAGWCPCTCPGGD---PHAGHTETSVLLHLSPAD
MMAR_1023|M.marinum_M EG-------------RDAGWSACGSAGGD---AHAGHTETSVLLHISPEV
MUL_0775|M.ulcerans_Agy99 EG-------------RDAGWSACGSAGGD---AHAGHTETSVLLHISPEV
MAV_4476|M.avium_104 EG-------------RDAGWCPCVAAGGD---AHAGHTETSVLLHISPAE
Mflv_5058|M.gilvum_PYR-GCK EG-------------RDAAWSPCVAGNAD---AHAGHTETSVLLHLSPDA
TH_1902|M.thermoresistible__bu EN-------------RDAAWLSCLAAGGD---AHAGHTETSVLLHLSPEV
MAB_3832c|M.abscessus_ATCC_199 EG-------------RDVAWWACAVEGGD---AHAGHVETSLLLHISPDV
Mvan_1602|M.vanbaalenii_PYR-1 EFPSLRTCFASLYDITPQARSGYFSDADD---PHANVAETSMLLHLAAGL
MSMEG_6870|M.smegmatis_MC2_155 HAEGTAAACSYWTLTAQTTAEAAATERPDRTPGHAGWFETSLMLAVHPEL
. * **. ***::* : .
Mb0714|M.bovis_AF2122/97 VRTERWRAGNRAP---LPVLLPSMRRGGVAAVSETG-VLGDPTTATAAEG
Rv0695|M.tuberculosis_H37Rv VRTERWRAGNRAP---LPVLLPSMRRGGVAAVSETG-VLGDPTTATAAEG
MMAR_1023|M.marinum_M VLTDRLSAGNAAP---LAELLPSLRRGGVAAVSPIG-VLGDPTTATAVEG
MUL_0775|M.ulcerans_Agy99 VLTDRLSAGNAAP---LAELLPSLRRGGVAAVSPIG-VLGDPTTATAVEG
MAV_4476|M.avium_104 VRTGQWQAGNRAP---LPELLGSMRRGGVAAVSPVG-VLGDPTTATAAEG
Mflv_5058|M.gilvum_PYR-GCK VRQEERVPGNRAP---LAELMPQMLRGGVAAVSSLG-VLGDPTTATADEG
TH_1902|M.thermoresistible__bu VSTEHVLPGNSAP---LAELMPQLRAGGVAAVSEVG-ILGDPTTATAAAG
MAB_3832c|M.abscessus_ATCC_199 VRREEVLAGNVAP---LHELMAPMRAGGVAAVSAIG-VLGDPTTASSQDG
Mvan_1602|M.vanbaalenii_PYR-1 VALDRAVDEEDRT---ARRVLTYAMP----DVTRSG-VVGRPTTADAGSG
MSMEG_6870|M.smegmatis_MC2_155 VADDRPFTPSDPPPLFNRSPAPGLTVERHGEWPRVGGTTDDAREASAETG
* . . . . * . . * : *
Mb0714|M.bovis_AF2122/97 RRIFAAMVDDCVRRVARWMPQPDGMLT--------------
Rv0695|M.tuberculosis_H37Rv RRIFAAMVDDCVRRVARWMPQPDGMLT--------------
MMAR_1023|M.marinum_M RRIFAEMVDDCVSRIARWTPGPDGMLT--------------
MUL_0775|M.ulcerans_Agy99 RRIFAEMVDDCVSRIARWTPGPDGMLT--------------
MAV_4476|M.avium_104 ERILSEMVNECVRRVARWRPGPDGMLA--------------
Mflv_5058|M.gilvum_PYR-GCK RRIFAEMVDACAARVVRWEPDRGGMLT--------------
TH_1902|M.thermoresistible__bu ARIFADMVDDCVERITRWTANRDGMLT--------------
MAB_3832c|M.abscessus_ATCC_199 ARMLETMTRQCADAVRRWTPDADGRLV--------------
Mvan_1602|M.vanbaalenii_PYR-1 ARLFDAIVDGVTHLLRQAREETEPMPEGPA-----------
MSMEG_6870|M.smegmatis_MC2_155 RLLLGRRVAGLVAALQSLDRATATPPAASVPSTDRERGPQA
:: . . :