For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TYLLNSPDDFADEAVRGLVAANPDLLTEVPGGVVRSTETPKGQPALVIGGGSGHYPAFAGWVGPGMGHGA PCGNIFSSPSASEVYSVVRNAENGGGVILGFGNYAGDVLHFGLAAEKLRHEGIDVRIVTVSDDIASNSPE NHRDRRGVAGDLPVFKIAGAAIEAGADLDEAERVAWKANDATRSFGLAFEGCTLPGATEPLFHVEKGWMG VGLGIHGEPGVRDNRLGTAAEVADMLFDEVTAEEPPRGENGYDGRVAVILNGLGTVKYEELFVVYGRIAE RLAQQGFTVVRPEVGEFVTSLDMAGVSLTMVFLDDELERLWTAPVETPAYRRGAMPAVDRTPRTTTWDAA ETTIPEASEGSRECARNIVAVLETFQQVCADNEAELGRIDAVAGDGDHGQGMSFGSRGAAQAARDAVDRN AGARTTLLLAGQAWADAAGGTSGALWGAALTSAGGVFSDTDGADEQAAVDAICAGIDAILRLGGAQPGDK TMVDAAVPFRDALVKAFDTQAGPAITSAARVAREAAEKTADITARRGRARVLGEKSVGTPDPGALSFAML MKALGEHLTR*
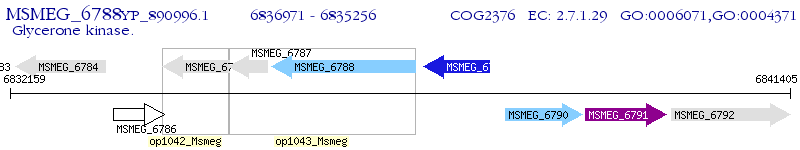
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6788 | - | - | 100% (571) | dihydroxyacetone kinase |
| M. smegmatis MC2 155 | MSMEG_3271 | - | e-152 | 50.78% (577) | dihydroxyacetone kinase |
| M. smegmatis MC2 155 | MSMEG_2123 | dhaK | 7e-61 | 39.40% (335) | dihydroxyacetone kinase, DhaK subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0194 | - | 5e-78 | 36.04% (566) | dihydroxyacetone kinase family protein |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1599 | - | 9e-05 | 27.68% (177) | NLP/P60 protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6788|M.smegmatis_MC2_155 ---MTYLLNSPDDFADEAVRGLVAANPDLLTEVPGG-VVRSTETPKGQPA
MAB_0194|M.abscessus_ATCC_1997 MVPLRNFLNTTESFIPDALRGLVAATADLWWDREHGYLIRDSPLRPGQVA
Mvan_1599|M.vanbaalenii_PYR-1 ---MPSAIVTALAAPIRRVQALVG--PGWSADPAADPAAALTGVRDTLTE
: : :. ::.**. .. : . :
MSMEG_6788|M.smegmatis_MC2_155 LVIGGGSGHYPAFAGWVGPGMGHGAPCGNIFSSPSASEVYSVVRNAENGG
MAB_0194|M.abscessus_ATCC_1997 IVSGGGSGHEPLHAGFIGRGMLTAACPGLIFTSPNALQIAGATVAADAGG
Mvan_1599|M.vanbaalenii_PYR-1 VADAAGRAWRDAEAGWSGSGGDAAAR----FAAATSAAIDGAAARAGQLG
:. ..* . **: * * .* *::..: : ... * *
MSMEG_6788|M.smegmatis_MC2_155 GVILGFGNYAGDVLHFGLAAEKLRHEGIDVRIVTVSDDIASNSPENHR-D
MAB_0194|M.abscessus_ATCC_1997 GVLHVVKNYTGDVMNFSIARQIAGESSVRTEVVLVDDDVATEVADAAKPG
Mvan_1599|M.vanbaalenii_PYR-1 ---------------VTAGDAAAAVAGAHRRLQSIVDEFEARAGALEPHL
. . . .: : *:. :.
MSMEG_6788|M.smegmatis_MC2_155 RRGVAGDLPVFKIAGAAIEAGADLDEAERVAWKANDATRSFGLAFEGCTL
MAB_0194|M.abscessus_ATCC_1997 RRGTAATVVVEKLCGAAAERGDQLAAVAALGRRVAASARSMAVALSPCTV
Mvan_1599|M.vanbaalenii_PYR-1 DSPGVARELLAEARDALRRAVAVVEELLAELDGHTAAAGAVAPAGSPAVA
.. : : .* . : :: :.. * . ..
MSMEG_6788|M.smegmatis_MC2_155 PGATEPLFHVEKGWMGVGLGIHGEPGVRDNRLGTAAEVADMLFDEVTAEE
MAB_0194|M.abscessus_ATCC_1997 PGAPVPSFDLPTGEMEIGVGIHGERGVDRAPVAPAAEIVAALLARILPAA
Mvan_1599|M.vanbaalenii_PYR-1 P-AGSPAVTAPAGWSGAGGAGPGPGGAPAGWAGPG---------------
* * * . * * . * *. ...
MSMEG_6788|M.smegmatis_MC2_155 PPRGENGYDGRVAVILNGLGTVKYEELFVVYGRIAERLAQQGFTVVRPEV
MAB_0194|M.abscessus_ATCC_1997 QVRSGD----EVIVVVNGLGATHSLELNLLFGEVAAQLAAAGIAVGRSLV
Mvan_1599|M.vanbaalenii_PYR-1 -------------AVNGGFGELAGFTPVEATGTPDAATFGDGVAVRLPDG
.: .*:* * *.:* .
MSMEG_6788|M.smegmatis_MC2_155 GEFVTSLDMAGVSLTMVFLDDELERLWTAPVETPAYRRGAMPAVDRTPRT
MAB_0194|M.abscessus_ATCC_1997 GSFVTALDMAGASITVVRADPEMFELWDAPTSAPGWPAVTGPPVGR----
Mvan_1599|M.vanbaalenii_PYR-1 TVVMAPNAVAASAVRHALTQLGVPYQWGGTTPGVGLDCSG----------
.::. :*. :: . : : * ... .
MSMEG_6788|M.smegmatis_MC2_155 TTWDAAETTIPEASEGSRECARNIVAVLETFQQVCADNEAELGRIDAVAG
MAB_0194|M.abscessus_ATCC_1997 ----LIDGTIVEPTDRADRGGEN--RWLSAFVERVRASVGMLTDLDRRAG
Mvan_1599|M.vanbaalenii_PYR-1 ---------------------------LTQWAYREAG-----LSLPRLAQ
* : : *
MSMEG_6788|M.smegmatis_MC2_155 DGDHGQGMSFGSRGAAQAARDAVDRNAGARTTLLLAGQAWADAAGGTSGA
MAB_0194|M.abscessus_ATCC_1997 DGDFGTNMAAALR-----HYELPLRGTDADVLLALS-TSYFVRAGGTSGA
Mvan_1599|M.vanbaalenii_PYR-1 EQDLGAAVG--------------------------------------AGS
: * * :. :*:
MSMEG_6788|M.smegmatis_MC2_155 LWGAALTSAGGVFSDTDGADEQAAVDAICAGIDAILRLGGAQPGDKTMVD
MAB_0194|M.abscessus_ATCC_1997 VFGTFFRALYGGLGSEPWSTERVAH-AVRHGLDRIQALGGARVGDKTVVD
Mvan_1599|M.vanbaalenii_PYR-1 LLPGDLAVWDGHVA--------------------------MIVGDGTMIE
: : * .. ** *:::
MSMEG_6788|M.smegmatis_MC2_155 AAVPFRDALVKAFDT--QAGPAITSAARVAREAAEKTADITARRGRARVL
MAB_0194|M.abscessus_ATCC_1997 AVSPAADALDAAVRQGIPLALALRSAADAAAAGVQATRDAIAARGRASYV
Mvan_1599|M.vanbaalenii_PYR-1 AGDPVKLSPIRTANA-----------------------------------
* * : :
MSMEG_6788|M.smegmatis_MC2_155 GEKSVGTPDPGALSFAMLMKALGEHLTR
MAB_0194|M.abscessus_ATCC_1997 GEAARGVEDPGALVMSWFFEAAAGR---
Mvan_1599|M.vanbaalenii_PYR-1 GQGFQGFWRPTA----------------
*: * * *