For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDDKLHGIPRVDLETRPRWKRDLAWWFGGKVLATARASAIWRKIAMPYEVPLIKATGGRARLSVGIPIAV LTSTGARSGKTRQTALAYFTDGDDVVLIASNYGQARHPGWYHNLRAHPECELYVGRRGGRFVAREVDGPQ RDRLYALAASRLYPGWVAYEKRAEGVRRIPVLRLTPADP
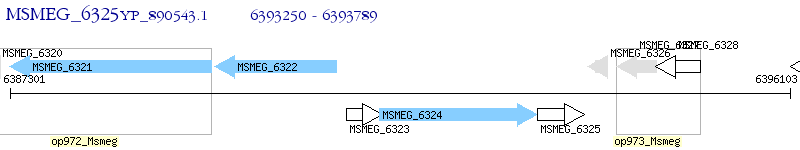
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6325 | - | - | 100% (179) | hypothetical protein MSMEG_6325 |
| M. smegmatis MC2 155 | MSMEG_3909 | - | 3e-40 | 48.33% (180) | hypothetical protein MSMEG_3909 |
| M. smegmatis MC2 155 | MSMEG_5215 | - | 8e-26 | 42.69% (171) | hypothetical protein MSMEG_5215 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1292c | - | 6e-15 | 35.16% (128) | hypothetical protein Mb1292c |
| M. gilvum PYR-GCK | Mflv_2085 | - | 2e-22 | 46.72% (137) | hypothetical protein Mflv_2085 |
| M. tuberculosis H37Rv | Rv1261c | - | 6e-15 | 35.16% (128) | hypothetical protein Rv1261c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1980c | - | 7e-17 | 39.82% (113) | hypothetical protein MAB_1980c |
| M. marinum M | MMAR_4178 | - | 6e-19 | 38.46% (130) | hypothetical protein MMAR_4178 |
| M. avium 104 | MAV_1448 | - | 1e-23 | 41.89% (148) | hypothetical protein MAV_1448 |
| M. thermoresistible (build 8) | TH_0359 | - | 2e-16 | 40.00% (110) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4480 | - | 1e-17 | 36.92% (130) | hypothetical protein MUL_4480 |
| M. vanbaalenii PYR-1 | Mvan_4618 | - | 1e-24 | 46.04% (139) | hypothetical protein Mvan_4618 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6325|M.smegmatis_MC2_155 MDDKLHGIPRVDLETRPRWKRDLAWWFGGKVLATARASAIWRKIAMPYEV
MAV_1448|M.avium_104 -------MPGRSFDDANAFHRLMRRFAS-----TTAGVALLRPTAHHLDR
Mflv_2085|M.gilvum_PYR-GCK -------MPLRHVDPHR--RRGHRYDQGVRFGRSKFGQFMARHIARRTDP
Mvan_4618|M.vanbaalenii_PYR-1 -------MPLRYVDPHR--RRGSRYQQGVRFGRSRVGQFIARHIARRTDP
Mb1292c|M.bovis_AF2122/97 ------------------------------MDISRWLERHVGVQLLRLHD
Rv1261c|M.tuberculosis_H37Rv ------------------------------MDISRWLERHVGVQLLRLHD
MMAR_4178|M.marinum_M ------------------------------MSNLTPFEQYVGIPILRLHD
MUL_4480|M.ulcerans_Agy99 ------------------------------MSNLTPFEQYVGIPILRLHD
MAB_1980c|M.abscessus_ATCC_199 ------------------------------------MSRLV-AKVLGVHQ
TH_0359|M.thermoresistible__bu --------------------------VSARGIRNSRWLASALKHFARGHI
.
MSMEG_6325|M.smegmatis_MC2_155 PLIKATGGR---ARLSVG-IPIAVLTSTGARSGKTRQTALAYFTDGDDVV
MAV_1448|M.avium_104 LVAKATGGRSSFAGLATG-IPVVMLTTTGAKSGEPRTVPVYGIPHPEGLA
Mflv_2085|M.gilvum_PYR-GCK ILFRLSKGR-----INMGPIVNAPLRTTGAKSGKPREVQLTYFHDGSDVI
Mvan_4618|M.vanbaalenii_PYR-1 MLFRLTGGR-----VNMGPIVNAPLRTVGAKSGKPREVQLTYFHDGHDVI
Mb1292c|M.bovis_AF2122/97 AIYRGTNGRIG--HRIPGAPPSLLLHTTGAKTSQPRTTSLTYARDGDAYL
Rv1261c|M.tuberculosis_H37Rv AIYRGTNGRIG--HRIPGAPPSLLLHTTGAKTSQPRTTSLTYARDGDAYL
MMAR_4178|M.marinum_M TIYRKTNGRIG--HRIPGVPPSLLLHTVGAKTGQARTTSLTYARDGDSYL
MUL_4480|M.ulcerans_Agy99 TIYRKTNGRIG--HRIPGVPPSLLLHTVGAKTGQARTTSLTSARDGDSYL
MAB_1980c|M.abscessus_ATCC_199 RIYIASKGYLG--HHVPGMAPNLLLTSVGAKSGARRINSLTYAHDGKDLV
TH_0359|M.thermoresistible__bu RVYRRSNGVLG--SRLLWFP-AALLTTIGRRTGEPRTTPTLYLRDGDRVI
: : * * : * ::. * .
MSMEG_6325|M.smegmatis_MC2_155 LIASNYGQARHPGWYHNLRAHPECELYVGRRGGRFVAREVDGPQRD--RL
MAV_1448|M.avium_104 LIASNFGGAKHPAWYFNLKAHPEATVTVGRDSWPATARLATPDERD--QI
Mflv_2085|M.gilvum_PYR-GCK LVASNFGGSSHPQWYHNLKANPECTFGQE----PFTAAEVTDAEEH--RR
Mvan_4618|M.vanbaalenii_PYR-1 LVASNFGGSTHPQWYYNLKAHPECDFGRE----PFTAEQVTDPGEY--SR
Mb1292c|M.bovis_AF2122/97 IVASKGGDPRSPGWYHNLKANPDVEINVGPKRFGVTAKPVQPHDPDYARL
Rv1261c|M.tuberculosis_H37Rv IVASKGGDPRSPGWYHNLKANPDVEINVGPKRFGVTAKPVQPHDPDYARL
MMAR_4178|M.marinum_M IVASNGGDERYPGWYHNLRKRPDCEINVGPKRIAVTARRVTPDDADYQRM
MUL_4480|M.ulcerans_Agy99 IVASNGGDERYPGWYHNLRKRPDCKINVGPKRIAVTARRVTPDDADYQRM
MAB_1980c|M.abscessus_ATCC_199 IVASNGGARRNPAWYHNLKAHPDVQVQLGREKRRVHATALLPGDADYERL
TH_0359|M.thermoresistible__bu LPASFGGRSEHPQWYRNLRANPRVHVQIGARHLDLVARDATAAERD--RY
: ** * * ** **: .* . *
MSMEG_6325|M.smegmatis_MC2_155 YALAASRLYPGWVAY-EKRAEGVRRIPVLRLTPADP
MAV_1448|M.avium_104 WAKG-VELYPGWRKY-EVRAG-DRHIEAFILSRAPA
Mflv_2085|M.gilvum_PYR-GCK LYELAERVYAGYRDYREKTSVTGRDIPVFRLTPR--
Mvan_4618|M.vanbaalenii_PYR-1 LYQLAERVYEGYADYRDKTADSGRQIPIFRLKPR--
Mb1292c|M.bovis_AF2122/97 WQIVNENNANRYTNY---QSRTSRPIPVVVLTRR--
Rv1261c|M.tuberculosis_H37Rv WQIVNENNANRYTNY---QSRTSRPIPVVVLTRR--
MMAR_4178|M.marinum_M WQLVNKNNANRYDSY---QRRTSRPIPIFALTPAA-
MUL_4480|M.ulcerans_Agy99 WQLVNKNNANRYDSY---QRRTSRPIPIFALTPAA-
MAB_1980c|M.abscessus_ATCC_199 WKLADTNNSGHYSAY---QRATSRPIPIVVLAPA--
TH_0359|M.thermoresistible__bu WPELTRIYP-PYRGY---RDATDRVIPLVVCEP---
: * * * .