For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSGNHDHDHDHARTVKPMVDEVTDFEVLEIALRELCIEKGIFTAEEHRRFTEFAEQIGPTPAARLVAKAW LDPEFKQLALTDALAASKAVGVDWLEPTGFGTPSDFTAFKILEDTPTLHHVIVCALCSCYPRPILGNSPE WYRTPNYRRRLVRWPRQVLAEFGLYLPDDVEVRVEDSNQKHRFMVMPMRPEGTEGWTEDQLAEIITRDCL IGVALPKPGVTTNVIVDTRPAIHPAG
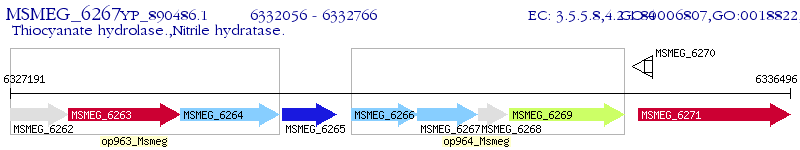
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6267 | - | - | 100% (236) | thiocyanate hydrolase gamma subunit |
| M. smegmatis MC2 155 | MSMEG_0378 | nthA | 7e-35 | 39.25% (186) | nitrile hydratase, alpha subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4916 | - | 1e-123 | 86.15% (231) | thiocyanate hydrolase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1504 | - | 1e-128 | 89.32% (234) | thiocyanate hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6267|M.smegmatis_MC2_155 MSG----NHDHDHDHARTVKPMVDEVTDFEVLEIALRELCIEKGIFTAEE
Mvan_1504|M.vanbaalenii_PYR-1 MTH----DHDHDHDHDRTVKPMVDEVTDFEVLEIALRELCIEKGIFTAED
Mflv_4916|M.gilvum_PYR-GCK MSHDHEHDHDHDHDHDRTVKPMVDEITDFEVLEIALRELCIEKGIFTAEE
*: :******* *********:***********************:
MSMEG_6267|M.smegmatis_MC2_155 HRRFTEFAEQIGPTPAARLVAKAWLDPEFKQLALTDALAASKAVGVDWLE
Mvan_1504|M.vanbaalenii_PYR-1 HRRFTEFSEQIGPTPAANLVARAWLDPDFKQLALTEPMTASKEVGVDWLE
Mflv_4916|M.gilvum_PYR-GCK HRRFTEYAEQIGPTPAATLVARAWLDPDFAELARTEPMTASKEVGVDWME
******::********* ***:*****:* :** *:.::*** *****:*
MSMEG_6267|M.smegmatis_MC2_155 PTGFGTPSDFTAFKILEDTPTLHHVIVCALCSCYPRPILGNSPEWYRTPN
Mvan_1504|M.vanbaalenii_PYR-1 PTGFGTPSDFTAFEILEDTATLHHVIVCALCSCYPRPILGNSPEWYRTPN
Mflv_4916|M.gilvum_PYR-GCK PTGFGTPSDFTAFEILADTPTLHNVIVCALCSCYPRPILGNSPEWYRTPN
*************:** **.***:**************************
MSMEG_6267|M.smegmatis_MC2_155 YRRRLVRWPRQVLAEFGLYLPDDVEVRVEDSNQKHRFMVMPMRPEGTEGW
Mvan_1504|M.vanbaalenii_PYR-1 YRRRLVRWPRQVLAEFGLYLPDDIEVRVQDSNQKHRYMVMPLRPEGTDGW
Mflv_4916|M.gilvum_PYR-GCK YRRRMVRWPRQVLSEFGLSLPEGVDVRVQDSNQKHRYMVMPMRPAGTDGW
****:********:**** **:.::***:*******:****:** **:**
MSMEG_6267|M.smegmatis_MC2_155 TEDQLAEIITRDCLIGVALPKPGVTTNVIVDTRPAIHPAG
Mvan_1504|M.vanbaalenii_PYR-1 TEQQLAEIVTRDCLIGVALPKPGVTSNVIVDTRPAVHP--
Mflv_4916|M.gilvum_PYR-GCK SEEQLAEIITRDCLIGVALPKPGVTTNVIVDTRPAVHP--
:*:*****:****************:*********:**