For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGRLKAHFPEIPDAPPPDLLDHDRFLAYMKTVHDVGGEPDAPMKYENKEYEYWEHMTYVICEVLAWRGIW LSEERRRIGNVDVGRAIYQGIPYYGRWLWAVARVLIEKHHISHGELTDRMVEVQERYRDGLAGRMLEATP KSEGDGADVRRNEHHIRAVGIGDPQIYAGKAGTPKFSVGEAVVVRDLPALLYTRTPEYVRGAKGVIAEVT YESPAPEDETWDRTDATPEWFYIVRFNLSELFYGYTGTSTDTLQTEIPERWLEKSL
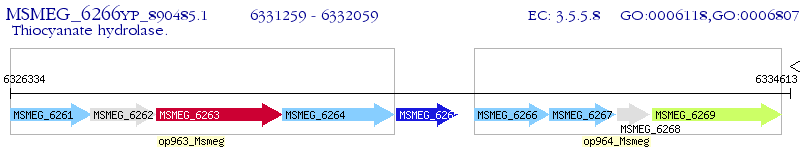
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6266 | - | - | 100% (266) | thiocyanate hydrolase beta subunit |
| M. smegmatis MC2 155 | MSMEG_0377 | - | 2e-05 | 22.94% (170) | nitrile hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4915 | - | 1e-122 | 76.43% (263) | hypothetical protein Mflv_4915 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1505 | - | 1e-122 | 77.48% (262) | hypothetical protein Mvan_1505 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4915|M.gilvum_PYR-GCK MSTAAERAAQLDLVARLKSAFPELPDAPTPDLLDHARITAYLKPVHDVGG
Mvan_1505|M.vanbaalenii_PYR-1 MSTAAERAAQLELVARLKSAFPELPDAPTPDLLDHPRITAYLKPVHDVGG
MSMEG_6266|M.smegmatis_MC2_155 -------------MGRLKAHFPEIPDAPPPDLLDHDRFLAYMKTVHDVGG
:.***: ***:****.****** *: **:*.******
Mflv_4915|M.gilvum_PYR-GCK EPDAPMKYENKQYEQWEEMTYVICEVLAWRGIWLSEERRRIGNVDVGRAE
Mvan_1505|M.vanbaalenii_PYR-1 EPDAPMKYENKQYEQWEELTYVICEVLAWRGIWLSEERRRIGNVDVGRAE
MSMEG_6266|M.smegmatis_MC2_155 EPDAPMKYENKEYEYWEHMTYVICEVLAWRGIWLSEERRRIGNVDVGRAI
***********:** **.:******************************
Mflv_4915|M.gilvum_PYR-GCK YLGLPYYGRWLLSVARVLVEKHHIGLTELSERMAEVAARYPDGLAGRKLE
Mvan_1505|M.vanbaalenii_PYR-1 YLGLPYYGRWLLAVARVLVEKHHIALGELSERMAEVQARYADGLNGRKLE
MSMEG_6266|M.smegmatis_MC2_155 YQGIPYYGRWLWAVARVLIEKHHISHGELTDRMVEVQERYRDGLAGRMLE
* *:******* :*****:*****. **::**.** ** *** ** **
Mflv_4915|M.gilvum_PYR-GCK AQPKSVGDGADVRRNAHHLHAVGKGDPQVYAGRAGEPRFAVGDAVVVREL
Mvan_1505|M.vanbaalenii_PYR-1 AQPKSAGDGSDVKRNAHHKRAVGKGDPQIYAGQAGEAKFKVGDRVLVRDL
MSMEG_6266|M.smegmatis_MC2_155 ATPKSEGDGADVRRNEHHIRAVGIGDPQIYAGKAGTPKFSVGEAVVVRDL
* *** ***:**:** ** :*** ****:***:** .:* **: *:**:*
Mflv_4915|M.gilvum_PYR-GCK PVLFYTRTPEYVRGAVGEIDTVAYESPAAEDETWDRPDATPEWFYIVKFM
Mvan_1505|M.vanbaalenii_PYR-1 PVLFYTRTPEYVRGATGEIDVVAYESPAAEDETWDRPDAKPEWFYIVKFA
MSMEG_6266|M.smegmatis_MC2_155 PALLYTRTPEYVRGAKGVIAEVTYESPAPEDETWDRTDATPEWFYIVRFN
*.*:*********** * * *:*****.*******.**.*******:*
Mflv_4915|M.gilvum_PYR-GCK MADLWHGYTGPSTDILRTEIPQRWLEAAH
Mvan_1505|M.vanbaalenii_PYR-1 MSDLWHGYTGTSTDTLRTEIPERWLAAAT
MSMEG_6266|M.smegmatis_MC2_155 LSELFYGYTGTSTDTLQTEIPERWLEKSL
:::*::****.*** *:****:*** :