For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDQVTVSLTEFDLRTTPLREVNAALHQPGLEGEFVIEHPAGAHNVAVGVDAPVRVRVEGHVGYYAAGMNQ QAGILINGNAGTGVAENMMSGTVRVKGNASQSAGATAHGGLLVVEGDAAARCGISMKGIDIVVGGNIGHM SAFMAQAGRLVVRGNAGEALGDSIYEARIYVRGEVASLGADCIAKPMRPEHHAELAELLAAAGYDDDTSE YARYGSARNLYHFHVDNASAY
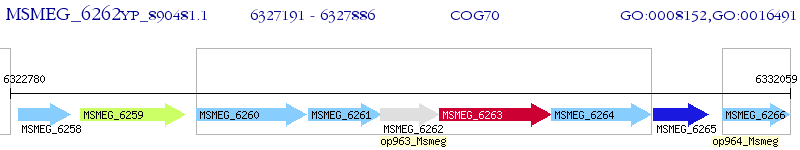
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6262 | - | - | 100% (231) | FwdC/FmdC family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1298 | - | 1e-104 | 82.43% (222) | glutamate synthase alpha subunit |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0608 | - | 5e-06 | 35.51% (107) | PE-PGRS family protein |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5511 | - | 1e-105 | 81.98% (222) | glutamate synthase alpha subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1298|M.gilvum_PYR-GCK ----MAALR-YNLREIPLREVNSALHAP----------------------
Mvan_5511|M.vanbaalenii_PYR-1 ----MEQMRTFDLRTTPLREVNTALHAA----------------------
MSMEG_6262|M.smegmatis_MC2_155 MDQVTVSLTEFDLRTTPLREVNAALHQP----------------------
MMAR_0608|M.marinum_M MSFVITAPEALTAAATDLASLGSTIGAANAAAVAPTTAVLAAGADEVSAA
* .:.::: .
Mflv_1298|M.gilvum_PYR-GCK --------------------DLDGEFVIENPAGAHNVAVGLNAPVTVTIA
Mvan_5511|M.vanbaalenii_PYR-1 --------------------DLTGDFLIEHPAGAHNVAVGLSAPVKVTID
MSMEG_6262|M.smegmatis_MC2_155 --------------------GLEGEFVIEHPAGAHNVAVGVDAPVRVRVE
MMAR_0608|M.marinum_M VAALFDAQGQAYQLLSARAATFHEQFLQALSGGGASYAAAEAAAVSPLLD
: :*: ..*. . *.. *.* :
Mflv_1298|M.gilvum_PYR-GCK GHVG---------YYAAGMNQQAEVIIDGNAGTGVAENMMSGSVWVKGNA
Mvan_5511|M.vanbaalenii_PYR-1 GHVG---------YYAAGMNQHADVIINGNAGTGVAENMMSGSVWVKGDA
MSMEG_6262|M.smegmatis_MC2_155 GHVG---------YYAAGMNQQAGILINGNAGTGVAENMMSGTVRVKGNA
MMAR_0608|M.marinum_M AINAPVLAATGRPLIGNGANGAPGTGADGGAAGWLIGNGGAGGSGTPGTA
. . . * * . :*.*. : * :* . * *
Mflv_1298|M.gilvum_PYR-GCK SQSAGATAHG---GLLVVEGDAAARCGISMKGVDIVVGGN----------
Mvan_5511|M.vanbaalenii_PYR-1 SQSAGATAHG---GLLVIEGNAAARCGISMKGVDIVVGGD----------
MSMEG_6262|M.smegmatis_MC2_155 SQSAGATAHG---GLLVVEGDAAARCGISMKGIDIVVGGN----------
MMAR_0608|M.marinum_M SVAGGNGGNGGAGGLLFGTGGAGGAGGTASSLVGAIPGGNGGYGGDGGLL
* :.* .:* ***. *.*.. * : . :. : **:
Mflv_1298|M.gilvum_PYR-GCK -------------------------------------VGHMSAFMAQAG-
Mvan_5511|M.vanbaalenii_PYR-1 -------------------------------------VGHMSAFMAQAG-
MSMEG_6262|M.smegmatis_MC2_155 -------------------------------------IGHMSAFMAQAG-
MMAR_0608|M.marinum_M FGRGGAGGAGGFGNNTTGGIGGLGGAGGLFGAGGDGGVGGFGTFAGNGGD
:* :.:* .:.*
Mflv_1298|M.gilvum_PYR-GCK ---RLVIRGDAGEALGDSIYEAR----------LYVRGEVASLG------
Mvan_5511|M.vanbaalenii_PYR-1 ---RLVVRGDAGEALGDSIYEAR----------IYVRGDVASLG------
MSMEG_6262|M.smegmatis_MC2_155 ---RLVVRGNAGEALGDSIYEAR----------IYVRGEVASLG------
MMAR_0608|M.marinum_M GGTGLFAAGGDGGAGGSGLTQGGDGGIGGAALGLFGAGGAGGTGGEGGLF
*. *. * * *..: :. :: * ... *
Mflv_1298|M.gilvum_PYR-GCK ------------------------ADCIAKPMRDEHHQELEYLLKTA---
Mvan_5511|M.vanbaalenii_PYR-1 ------------------------ADCVAKPMRDEHHSELEKLLKAA---
MSMEG_6262|M.smegmatis_MC2_155 ------------------------ADCIAKPMRPEHHAELAELLAAA---
MMAR_0608|M.marinum_M GGAGGMGGSAGMLFGNGGDGGAGAAATIGNAAGNGGAGGNAGMLIGAGGA
* :.:. :* *
Mflv_1298|M.gilvum_PYR-GCK ------GFDDDDTKSYTRYGSARSLYHFHVDNASSY--------------
Mvan_5511|M.vanbaalenii_PYR-1 ------GFEGDDTRSYTRYGSARSLYHFHVDNASAY--------------
MSMEG_6262|M.smegmatis_MC2_155 ------GYD-DDTSEYARYGSARNLYHFHVDNASAY--------------
MMAR_0608|M.marinum_M GGNGGFGFSASDGGAGGAGGNAGFLYGAGGAGGAGGDSVGGADGGDGGNG
*:. .* *.* ** ..:.
Mflv_1298|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5511|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6262|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0608|M.marinum_M GKAGLVGQGGDGGAGGNNNSGFGTGGSGGKGGDAVLIGNGGNGGNAGSGG
Mflv_1298|M.gilvum_PYR-GCK --------------------------
Mvan_5511|M.vanbaalenii_PYR-1 --------------------------
MSMEG_6262|M.smegmatis_MC2_155 --------------------------
MMAR_0608|M.marinum_M VGPGIAGAAGIGGLLIGEDGMAGLTP