For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DVAVESTEMYPGDIKSVLLSEEQIRTRTAELAAAIADQYRDNLGDDDLLLVTVLKGAVMFVTDLARSIPL PTQLEFMAVSSYGSSTSSSGVVRILKDLDRDINDRDVLIVEDIIDSGLTLSWLLRNLATRHPRSLKVCTL LRKPEAVRTDIDVEYVGFDIPNEFVVGYGLDYAERYRDLPFIGLLDPKVYTH*
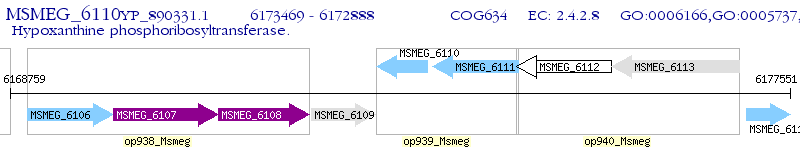
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6110 | hpt | - | 100% (193) | hypoxanthine phosphoribosyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3648c | hpt | 5e-76 | 72.87% (188) | hypoxanthine-guanine phosphoribosyltransferase |
| M. gilvum PYR-GCK | Mflv_1412 | - | 2e-87 | 83.60% (189) | hypoxanthine phosphoribosyltransferase |
| M. tuberculosis H37Rv | Rv3624c | hpt | 5e-76 | 72.87% (188) | hypoxanthine-guanine phosphoribosyltransferase |
| M. leprae Br4923 | MLBr_00214 | hpt | 6e-80 | 76.47% (187) | putative hypoxanthine phosphoribosyltransferase |
| M. abscessus ATCC 19977 | MAB_0523 | - | 2e-77 | 78.26% (184) | hypoxanthine phosphoribosyltransferase (HPT) |
| M. marinum M | MMAR_5124 | hpt | 1e-77 | 76.76% (185) | hypoxanthine-guanine phosphoribosyltransferase Hpt |
| M. avium 104 | MAV_0532 | hpt | 1e-78 | 76.06% (188) | hypoxanthine phosphoribosyltransferase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4200 | hpt | 9e-78 | 76.76% (185) | hypoxanthine-guanine phosphoribosyltransferase Hpt |
| M. vanbaalenii PYR-1 | Mvan_5377 | - | 1e-88 | 83.77% (191) | hypoxanthine phosphoribosyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6110|M.smegmatis_MC2_155 ----------------------MDVAVESTEMYPGDIKSVLLSEEQIRTR
MAB_0523|M.abscessus_ATCC_1997 --------------------MTCEQDQACGEQYSGDVKSVLLTEEQIRAK
Mb3648c|M.bovis_AF2122/97 MTPALVVGPAAWHAVHVTQSSSAITPGQTAELYPGDIKSVLLTAEQIQAR
Rv3624c|M.tuberculosis_H37Rv MTPALVVGPAAWHAVHVTQSSSAITPGQTAELYPGDIKSVLLTAEQIQAR
MMAR_5124|M.marinum_M --------------MHVAQSSSAITPGHAGELYPGDIKSVLLTEEQIQRR
MUL_4200|M.ulcerans_Agy99 --------------MHVAQSSSAITPGHTGELYPGDIKSVLLTEEQIQRR
MAV_0532|M.avium_104 --------------MGVAQISSAITPAQPAELYPGDIKSVLLTAEQIQAR
MLBr_00214|M.leprae_Br4923 ----MVFRRAAWHALLVAQSFSAINPGPTAELYLGDIKSVMLTQEQIQAR
Mflv_1412|M.gilvum_PYR-GCK ------------------------MPAHTADLYSGDIKSVLLSEDQIRAK
Mvan_5377|M.vanbaalenii_PYR-1 ----------------------MHVPTRTAELYPGDIKSVLLSEEQIKAK
: * **:***:*: :**: :
MSMEG_6110|M.smegmatis_MC2_155 TAELAAAIADQYRDNLGDD--DLLLVTVLKGAVMFVTDLARSIPLPTQLE
MAB_0523|M.abscessus_ATCC_1997 TQELGAAIGEKYRDIEG----DLLLVTVLKGAVMFVSDLARAIPIPTQME
Mb3648c|M.bovis_AF2122/97 IAELGEQIGNDYRELSATTGQDLLLITVLKGAVLFVTDLARAIPVPTQFE
Rv3624c|M.tuberculosis_H37Rv IAELGEQIGNDYRELSATTGQDLLLITVLKGAVLFVTDLARAIPVPTQFE
MMAR_5124|M.marinum_M IDELAAQIGNDYRELADTSDQDLLLITVLKGAVLFVTDLARAIPLPTQFE
MUL_4200|M.ulcerans_Agy99 IDELAAQIGNDYRELVDTSDQDLLLITVLKGAVLFVTDLARAIPLPTQFE
MAV_0532|M.avium_104 IAELGREIGDNYRDAAAETGQDLLLITVLKGAVIFVTDLARAIPLPTQFE
MLBr_00214|M.leprae_Br4923 IAELGAQIGHDYRDRSPASDQDLLLVTVLKGAVIFVTDLARAIPLPTQFE
Mflv_1412|M.gilvum_PYR-GCK TAELAALIADDYQDR--ESGQDLLLITVLKGAVMFVTDLARAIPLPTQLE
Mvan_5377|M.vanbaalenii_PYR-1 TAELAAQIAGDYQDA--ESGQDLLLVTVLKGAVMFVTDLARAIPVPTQLE
**. *. .*:: ****:*******:**:****:**:***:*
MSMEG_6110|M.smegmatis_MC2_155 FMAVSSYGSSTSSSGVVRILKDLDRDINDRDVLIVEDIIDSGLTLSWLLR
MAB_0523|M.abscessus_ATCC_1997 FMAVSSYGSATSSSGVVRILKDLDRDIQNLDVLIVEDIIDSGLTLSWLMR
Mb3648c|M.bovis_AF2122/97 FMAVSSYGSSTSSSGVVRILKDLDRDIHGRDVLIVEDVVDSGLTLSWLSR
Rv3624c|M.tuberculosis_H37Rv FMAVSSYGSSTSSSGVVRILKDLDRDIHGRDVLIVEDVVDSGLTLSWLSR
MMAR_5124|M.marinum_M FMAVSSYGSSTSSSGVVRILKDLDRDIAGRDVLIVEDVVDSGLTLSWLLR
MUL_4200|M.ulcerans_Agy99 FMAVSSYGSSTSSSGVVRILKDLDRDIAGRDVLIVEDVVDSGLTLSWLLR
MAV_0532|M.avium_104 FMAVSSYGSSTSSSGVVRILKDLDRDIQGRDVLIVEDVVDSGLTLSWLLR
MLBr_00214|M.leprae_Br4923 FMAVSSYGSSMSSSGVVRILKDLDRDIHDRNVLIVEDVVDSGLTLSWLLR
Mflv_1412|M.gilvum_PYR-GCK FMAVSSYGSSTSSSGVVRILKDLDRDINGRDVLIVEDIIDSGLTLSWLLR
Mvan_5377|M.vanbaalenii_PYR-1 FMAVSSYGSSTSSSGVVRILKDLDRDINDRDVLIVEDIVDSGLTLSWLLR
*********: **************** . :******::********* *
MSMEG_6110|M.smegmatis_MC2_155 NLATRHPRSLKVCTLLRKPEAVRTDIDVEYVGFDIPNEFVVGYGLDYAER
MAB_0523|M.abscessus_ATCC_1997 NLASRGPRSLNVVSLLRKPEAKKVDVDVALVGFEIPNEFVVGYGLDYGER
Mb3648c|M.bovis_AF2122/97 NLTSRNPRSLRVCTLLRKPDAVHANVEIAYVGFDIPNDFVVGYGLDYDER
Rv3624c|M.tuberculosis_H37Rv NLTSRNPRSLRVCTLLRKPDAVHANVEIAYVGFDIPNDFVVGYGLDYDER
MMAR_5124|M.marinum_M NLKSRQPKSLRVCTLLRKPEAVGANVDITYVGFDISNDFVVGYGLDYDER
MUL_4200|M.ulcerans_Agy99 NLKSRQPKSLRVCTLLRKPEAVGANVDITYVGFDISNDFVVGYGLDYDER
MAV_0532|M.avium_104 NLSTRHPRSLRVCTLLRKPDARGAHVDIAYVGFDIPNDFVVGYGLDYDER
MLBr_00214|M.leprae_Br4923 NLATRRPQSLQVCTLLRKPDALRANVDITYVGFDIPNDFVVGYGLDYDER
Mflv_1412|M.gilvum_PYR-GCK NLATRGPKSLRVCTLLRKPEAVRAELDVEYIGFDIPNEFVVGYGLDYAER
Mvan_5377|M.vanbaalenii_PYR-1 NLATRGPRSLRVCTLLRKPEAVRADVDISYIGFDIPNEFVVGYGLDYAER
** :* *:**.* :*****:* ..::: :**:*.*:********* **
MSMEG_6110|M.smegmatis_MC2_155 YRDLPFIGLLDPKVYTH-
MAB_0523|M.abscessus_ATCC_1997 YRDLPFIGTLHPRVYTD-
Mb3648c|M.bovis_AF2122/97 YRDLSYIGTLDPRVYQ--
Rv3624c|M.tuberculosis_H37Rv YRDLSYIGTLDPRVYQ--
MMAR_5124|M.marinum_M YRDLSYIGTLDPRVYQ--
MUL_4200|M.ulcerans_Agy99 YRDLSYIGTLDPRVYQ--
MAV_0532|M.avium_104 YRDLSYIGTLDPRVYQQ-
MLBr_00214|M.leprae_Br4923 YRDLPYIGMLDPRVYQQ-
Mflv_1412|M.gilvum_PYR-GCK YRDLPYIGTLDPKVYKP-
Mvan_5377|M.vanbaalenii_PYR-1 YRDLPYIGTLDPKVYEQT
****.:** *.*:**