For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPAPQPQPVLAPLTPAAVFLVATIDEGQEATVYDALPDISGLVRAIGFRDPAKRLSAITSIGSDAWDRLF SGPRPAELHPFREIDGGRHHAPATPGDLLFHLRAESMDVCFELATKLVEAMSGAITIVDETHGFRFFDNR DLMGFVDGTENPDGNLAVVATQIGDEDPDFAGGCYVHVQKYLHDMASWNSLSVEEQERVIGRTKLDDIEL DDDVKPANSHVALNVIEDEDGNELKIIRHNMPFGEIGKGEFGTYYIGYSRTPSVTERMLDNMFIGDPPGN TDRILDFSTAITGGLFFTPTVDFLDDPPPLPSEDDRAEPASAPSADPVHTDGSLGIGSLKGTR
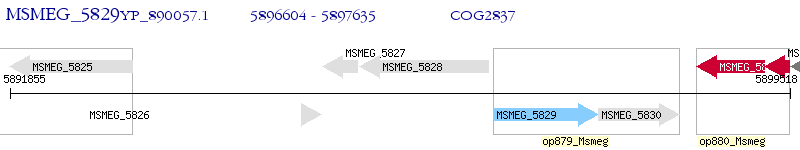
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5829 | - | - | 100% (343) | Dyp-type peroxidase |
| M. smegmatis MC2 155 | MSMEG_5420 | - | 9e-08 | 25.60% (250) | Tat-translocated enzyme |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0822c | - | 1e-151 | 73.98% (342) | hypothetical protein Mb0822c |
| M. gilvum PYR-GCK | Mflv_1627 | - | 1e-162 | 79.94% (344) | Dyp-type peroxidase family protein |
| M. tuberculosis H37Rv | Rv0799c | - | 1e-151 | 73.98% (342) | hypothetical protein Rv0799c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0700c | - | 1e-141 | 69.01% (342) | putative Dyp-type peroxidase |
| M. marinum M | MMAR_4892 | - | 1e-156 | 75.66% (341) | hypothetical protein MMAR_4892 |
| M. avium 104 | MAV_0741 | - | 1e-156 | 76.32% (342) | Dyp-type peroxidase family protein |
| M. thermoresistible (build 8) | TH_0569 | - | 1e-159 | 76.61% (342) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0406 | - | 1e-156 | 75.44% (342) | hypothetical protein MUL_0406 |
| M. vanbaalenii PYR-1 | Mvan_5122 | - | 1e-163 | 81.23% (341) | Dyp-type peroxidase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1627|M.gilvum_PYR-GCK --MPAPLPQPVLAPLTPAAIFLVATIDDGGERAVHDALPDIAGLVRAVGF
Mvan_5122|M.vanbaalenii_PYR-1 --MPAPLPQPVLAPLTPAAIFLVATIDEGGEEAVHDALGDIAGLVRAVGF
MSMEG_5829|M.smegmatis_MC2_155 --MPAPQPQPVLAPLTPAAVFLVATIDEGQEATVYDALPDISGLVRAIGF
Mb0822c|M.bovis_AF2122/97 MAVPAVSPQPILAPLTPAAIFLVATIGADGEATVHDALSKISGLVRAIGF
Rv0799c|M.tuberculosis_H37Rv MAVPAVSPQPILAPLTPAAIFLVATIGADGEATVHDALSKISGLVRAIGF
MMAR_4892|M.marinum_M --MPSVQPQPILAPLTPAAIFLVATIDAGGEQTVHDALTEISGLVRAIGF
MUL_0406|M.ulcerans_Agy99 --MPSVQPQPILAPLTPAAIFLVATIDAGGEQTVHDALTEISGLVRAIGF
MAV_0741|M.avium_104 --MPPVQPQPILAPLTPAAIFLVLTVDDGGEATVHEALQDISGLVRAIGF
TH_0569|M.thermoresistible__bu --MAEVLPQPILEPLTPAAIFLVATIDKGGAARVHDVLPELSGMVRAIGF
MAB_0700c|M.abscessus_ATCC_199 --MPDSVPQAVLEPLSSSAIFLVVTIDEGAEQAVHGALPELSGLARAIGF
:. **.:* **:.:*:*** *:. . *: .* .::*:.**:**
Mflv_1627|M.gilvum_PYR-GCK RDPAKRLSVVTSIGSDAWDRLFSGPRPAELHPFVELDGGRHRAPSTPGDL
Mvan_5122|M.vanbaalenii_PYR-1 RDPAKRLSVVTSIGSRAWDRLFSGPRPAELHPFIELDGGRHHAPSTPGDL
MSMEG_5829|M.smegmatis_MC2_155 RDPAKRLSAITSIGSDAWDRLFSGPRPAELHPFREIDGGRHHAPATPGDL
Mb0822c|M.bovis_AF2122/97 RDPTKHLSVVVSIGSDAWDRLFAGPRPTELHPFVELTGPRHTAPATPGDL
Rv0799c|M.tuberculosis_H37Rv RDPTKHLSVVVSIGSDAWDRLFAGPRPTELHPFVELTGPRHTAPATPGDL
MMAR_4892|M.marinum_M RDPTKHLSVVASIGSDAWDRLFSGPKPAELHPFVPLTGPRHNAPATPGDL
MUL_0406|M.ulcerans_Agy99 RDPTKHLSVVASIGADAWDRLFSGPKPAELHPFVPLTGPRHNAPATPGDL
MAV_0741|M.avium_104 REPQKRLSAIASIGSDVWDRLFSGLRPAELHRFVELHGPRHTAPATPGDL
TH_0569|M.thermoresistible__bu RDPTKRLSMVTSIGSDAWDQLFSGPRPAELHPFIELEGPRHKAPATPGDL
MAB_0700c|M.abscessus_ATCC_199 RDPARRLSLVTAIGSEAWDRLFSGSRPARLWPFREVRGERHVAPATPGDL
*:* ::** :.:**: .**:**:* :*:.* * : * ** **:*****
Mflv_1627|M.gilvum_PYR-GCK LFHIRAETMDVCFELATKLVAAMG-AITVVDETHGFRFFDNRDLLGFVDG
Mvan_5122|M.vanbaalenii_PYR-1 LFHIRAETMDVCFELATKLVAAMG-AITVVDETHGFRFFDNRDLLGFVDG
MSMEG_5829|M.smegmatis_MC2_155 LFHLRAESMDVCFELATKLVEAMSGAITIVDETHGFRFFDNRDLMGFVDG
Mb0822c|M.bovis_AF2122/97 LFHIRAETMDVCFELAGRILKSMGDAVTVVDEVHGFRFFDNRDLLGFVDG
Rv0799c|M.tuberculosis_H37Rv LFHIRAETMDVCFELAGRILKSMGDAVTVVDEVHGFRFFDNRDLLGFVDG
MMAR_4892|M.marinum_M LLHIRAETMDVCFELAGRILKAMAPAVTVVDEVHGFRFFDNRDLLGFVDG
MUL_0406|M.ulcerans_Agy99 LLHIRAETMDVCFELAGRILKAMAPAVTVVDEVHGFRFFDNRDLLGFVDG
MAV_0741|M.avium_104 LFHIRAESLDVCFELADRILKSMAGAVTVVDEVHGFRYFDNRDLLGFVDG
TH_0569|M.thermoresistible__bu LFHIRAEALDVCFELAGRIVKAMDGAITVVDEVHGFRFFDNRDLLGFVDG
MAB_0700c|M.abscessus_ATCC_199 LFHIRASTMDRCFELAARITTMLAGAVTVADEVHGFQYFDNRDLLGFVDG
*:*:**.::* ***** :: : *:*:.**.***::******:*****
Mflv_1627|M.gilvum_PYR-GCK TENPDGPLADSATRIGDEDPDFAGGCYVHVQKYVHRMDAWNALSTEEQER
Mvan_5122|M.vanbaalenii_PYR-1 TENPDGPLADSATQIGDEDPDFAGGCYVHVQKYLHLMDAWNALSTEEQER
MSMEG_5829|M.smegmatis_MC2_155 TENPDGNLAVVATQIGDEDPDFAGGCYVHVQKYLHDMASWNSLSVEEQER
Mb0822c|M.bovis_AF2122/97 TENPSGPIAIKATTIGDEDRNFAGSCYVHVQKYVHDMASWESLSVTEQER
Rv0799c|M.tuberculosis_H37Rv TENPSGPIAIKATTIGDEDRNFAGSCYVHVQKYVHDMASWESLSVTEQER
MMAR_4892|M.marinum_M TENPGGAIAIGATTVGDEDPDFAGSCYVHVQKYVHDMGSWDSLSVTEQER
MUL_0406|M.ulcerans_Agy99 TENPGGAIAIGATTVGDEDPDFAGSCYVHVQKYVHDMGSWDSLSVTEQER
MAV_0741|M.avium_104 TENPDGALAVSSTAIGDEDPDFAGSCYVHVQKYLHDMSAWTALSVTEQEN
TH_0569|M.thermoresistible__bu TENPDGPEARSATRIGDEDPDFEGGCYVHIQKYVHDIAAWEALSVDEQQR
MAB_0700c|M.abscessus_ATCC_199 TENPDGADAAAAALVGPEDPDFIGGSYVHVQKYLHNMAAWSALPVDEQEK
****.* * :: :* ** :* *..***:***:* : :* :*.. **:.
Mflv_1627|M.gilvum_PYR-GCK VIGRTKLDDIELDDAVKPADSHVALNVITDEDGTELKIVRHNMPFGEVGK
Mvan_5122|M.vanbaalenii_PYR-1 VIGRTKLDDIELDDAVKPANSHVALNVIEDEDGTELKIVRHNMPFGEIGA
MSMEG_5829|M.smegmatis_MC2_155 VIGRTKLDDIELDDDVKPANSHVALNVIEDEDGNELKIIRHNMPFGEIGK
Mb0822c|M.bovis_AF2122/97 VIGRTKLDDIELDDNAKPANSHVALNVITDDDGTERKIVRHNMPFGEVGK
Rv0799c|M.tuberculosis_H37Rv VIGRTKLDDIELDDNAKPANSHVALNVITDDDGTERKIVRHNMPFGEVGK
MMAR_4892|M.marinum_M VIGRTKLEDIELDESTKPANSHIALNVITDEDGVELKIVRHNMPFGEVGK
MUL_0406|M.ulcerans_Agy99 VIGRTKLEDIELDESTKPANSHIALNVITDEDGVELKIVRHNMPFGEVGK
MAV_0741|M.avium_104 VIGRTKLDDIELDDDVKPADAHIALNVITDDDGTELKIVRHNMPFGELGK
TH_0569|M.thermoresistible__bu VIGRTKLDDIELDDDVKPTNSHIALNVIEDEDGNELQIVRHNMPFGEVGK
MAB_0700c|M.abscessus_ATCC_199 VIGRTKLDDVEFDDAVKPSNSHVALNVIEDEEGNELQVLRRNMPFGELGK
*******:*:*:*: .**:::*:***** *::* * :::*:******:*
Mflv_1627|M.gilvum_PYR-GCK GEFGTYFIGYSRTAAVTERMLRNMFLGDPPGNTDRVLDFSTAVTGSMFFT
Mvan_5122|M.vanbaalenii_PYR-1 GEFGTYFIGYSRTAAVTERMLRNMFIGDPPGNTDRVLDFSTAVTGSMFFT
MSMEG_5829|M.smegmatis_MC2_155 GEFGTYYIGYSRTPSVTERMLDNMFIGDPPGNTDRILDFSTAITGGLFFT
Mb0822c|M.bovis_AF2122/97 GEYGTYFIGYSRTPTVTEQMLRNMFLGDPAGNTDRVLDFSTAVTGGLFFS
Rv0799c|M.tuberculosis_H37Rv GEYGTYFIGYSRTPTVTEQMLRNMFLGDPAGNTDRVLDFSTAVTGGLFFS
MMAR_4892|M.marinum_M GEYGTYFIGYSRTPVVTERLLRNMFLGDPPGNTDRVLDFSTATTGGLFFS
MUL_0406|M.ulcerans_Agy99 GEYGTYFIGYSRTPTVTERLLRNMFLGDSPGNTDRVLDFSTATTGGLFFS
MAV_0741|M.avium_104 SEYGTYFIGYSRTPRVTEQMLRNMFLGDPPGNTDRILDFSTAVTGGLFFS
TH_0569|M.thermoresistible__bu GEYGTYFIGYSRTPSVTEQMLRNMFLGDPPGNTDRILDFSTAVTGGLFFA
MAB_0700c|M.abscessus_ATCC_199 SEFGTYYIAYSRTPDVTERMLHNMFVGDPPGNTDRILDFSTAITGGLFFA
.*:***:*.****. ***::* ***:**..*****:****** **.:**:
Mflv_1627|M.gilvum_PYR-GCK PINDFLDDPPPLPGAVPDTDDTDDPDGSAPVG-----YHGSLAIGSLKVR
Mvan_5122|M.vanbaalenii_PYR-1 PIMDFLNNPPPLP---NSASDTVAPTDVTPVG-----YAGSLAIGSLKGQ
MSMEG_5829|M.smegmatis_MC2_155 PTVDFLDDPPPLPSEDDRAEPASAPS-ADPVH-----TDGSLGIGSLKGT
Mb0822c|M.bovis_AF2122/97 PTIDFLDHPPPLP---QAATPTLAA--------------GSLSIGSLKGS
Rv0799c|M.tuberculosis_H37Rv PTIDFLDHPPPLP---QAATPTLAA--------------GSLSIGSLKGS
MMAR_4892|M.marinum_M PTIDFLDDPPPLPP-APAAVPTTDD--------------GSLSIGSLKGI
MUL_0406|M.ulcerans_Agy99 PTIDFLDDPPPLPL-APAAVPTADD--------------GSLSIGSLKGT
MAV_0741|M.avium_104 PTVDFLDDPPPLPAPAMPAAPPARN--------------GSLSIGSLKGT
TH_0569|M.thermoresistible__bu PTVDFLNDPPPLPHTPAAAAQDVAETGDTSQTHDGEIHDGSLGIGSLKGI
MAB_0700c|M.abscessus_ATCC_199 PAADFLDAPPSLP--AEVAIKSVEYQ-------------GSLSIGSLKGS
* ***: **.** : ***.*****
Mflv_1627|M.gilvum_PYR-GCK RG
Mvan_5122|M.vanbaalenii_PYR-1 PQ
MSMEG_5829|M.smegmatis_MC2_155 R-
Mb0822c|M.bovis_AF2122/97 PR
Rv0799c|M.tuberculosis_H37Rv PR
MMAR_4892|M.marinum_M SE
MUL_0406|M.ulcerans_Agy99 SE
MAV_0741|M.avium_104 TR
TH_0569|M.thermoresistible__bu P-
MAB_0700c|M.abscessus_ATCC_199 LS