For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LVTLDGQLHDPDAPLLCADDLAAVRGDGVFETLLVRDGQPCLLEAHLARLTQSARMLDLPEPDLDAWRAA VALGVKRWTSEHSANGEEGVLRLVYSRGREGGGPPTAFATIGALADRVAGARRHGVAAITLDRGLPVGAS EMPWLAAGAKTLSYAVNMAALRHAARNGAGDVIFVSSDGFLLEGPRSTVVIATRGDDGQPLLLTPPPWYP ILRGTTQQALFETARNKGIDCDYRALRPADLLTAQGIWLISSITLAARVHTLDGTRLRTSASAPDIAGLV DTAILGDR*
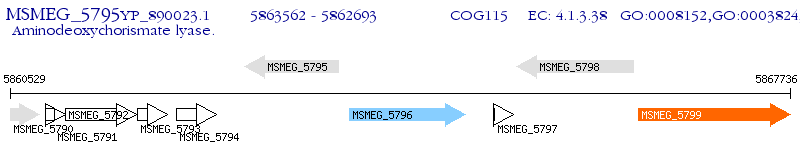
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5795 | - | - | 100% (289) | 4-amino-4-deoxychorismate lyase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0835 | - | 1e-112 | 70.21% (292) | 4-amino-4-deoxychorismate lyase |
| M. gilvum PYR-GCK | Mflv_1639 | - | 1e-114 | 71.38% (290) | 4-amino-4-deoxychorismate lyase |
| M. tuberculosis H37Rv | Rv0812 | - | 1e-112 | 70.21% (292) | 4-amino-4-deoxychorismate lyase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0731 | - | 2e-89 | 59.44% (286) | 4-amino-4-deoxychorismate lyase |
| M. marinum M | MMAR_4873 | pabC | 1e-109 | 68.60% (293) | amino acid aminotransferase, PabC |
| M. avium 104 | MAV_0751 | - | 1e-111 | 68.49% (292) | 4-amino-4-deoxychorismate lyase |
| M. thermoresistible (build 8) | TH_0072 | pabC | 1e-113 | 70.69% (290) | PROBABLE AMINO ACID AMINOTRANSFERASE |
| M. ulcerans Agy99 | MUL_0426 | pabC | 1e-108 | 67.92% (293) | 4-amino-4-deoxychorismate lyase |
| M. vanbaalenii PYR-1 | Mvan_5108 | - | 1e-114 | 70.69% (290) | 4-amino-4-deoxychorismate lyase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0835|M.bovis_AF2122/97 ------MVVTLDGEILQPGMPLLHADDLAAVRGDGVFETLLVRDGRACLV
Rv0812|M.tuberculosis_H37Rv ------MVVTLDGEILQPGMPLLHADDLAAVRGDGVFETLLVRDGRACLV
MMAR_4873|M.marinum_M MTGQTGVVVTLDGEIPEPGTPLLHADDLAAVRGDGVFETLLVRDGAPCLV
MUL_0426|M.ulcerans_Agy99 MTGQTGVVVTLDGEIPEPGTPLLHADDLAAVRGDGVFETLLVRDRAPCLV
MAV_0751|M.avium_104 ------MIVTLDGEVHAPDAPLLHADDLAAVRGDGVFETLLVRDGRACLI
TH_0072|M.thermoresistible__bu MANAPAVVVTLDGQLRAPGEPLLYADDLAAVRGDGIFETLLIRDGGPCLL
Mflv_1639|M.gilvum_PYR-GCK MADHPGVLVTLDGQLHDPEVPLLHADDLAAVRGDGIFETLLVRDGRPCLL
Mvan_5108|M.vanbaalenii_PYR-1 MADHPGVVVTLDGQLHDPDAPLLYADDLAAVRGDGIFETLLIRGGRPCLL
MSMEG_5795|M.smegmatis_MC2_155 ------MLVTLDGQLHDPDAPLLCADDLAAVRGDGVFETLLVRDGQPCLL
MAB_0731|M.abscessus_ATCC_1997 --MAVAVVVTLDGEVHDPSVPLLYADELAAVRGDGAFETALVRGGAVCKL
::*****:: * *** **:******** *** *:*. * :
Mb0835|M.bovis_AF2122/97 EAHLQRLTQSARLMDLPEPDLPRWRRAVEVATQRWVAST---ADEGALRL
Rv0812|M.tuberculosis_H37Rv EAHLQRLTQSARLMDLPEPDLPRWRRAVEVATQRWVAST---ADEGALRL
MMAR_4873|M.marinum_M EPHLQRLTTSARLMDLPAPDLARWRHAIDVAARRWVAST---GAEGSLRL
MUL_0426|M.ulcerans_Agy99 EPHLQRLTTSARLMDLPAPDLARWRHAIDVAARRWVAST---GAEGSLRL
MAV_0751|M.avium_104 ESHLQRLTQSAALTELPEPDLPRWRSAIEAATRRWCAGG---ADEGAMRL
TH_0072|M.thermoresistible__bu EAHLTRLTGSARLMDLPEPDLPAWRAAVATAVRQWCDRT---DDEGVMRL
Mflv_1639|M.gilvum_PYR-GCK DAHLGRLAHSAQMVDLPAPNAARWRTAVDTAVRAWLDSG---GDEGVLRL
Mvan_5108|M.vanbaalenii_PYR-1 DAHLGRLAHSARQVDLPPPDAARWHAAVGIAVDSWLRTG---GDEGVLRL
MSMEG_5795|M.smegmatis_MC2_155 EAHLARLTQSARMLDLPEPDLDAWRAAVALGVKRWTSEHSANGEEGVLRL
MAB_0731|M.abscessus_ATCC_1997 EAHLGRMAASAASMDLAEPDRAAWRSAVKIAVQQWNSIS---GEDAMLRL
:.** *:: ** :*. *: *: *: .. * :. :**
Mb0835|M.bovis_AF2122/97 IYSRGREGGS-APTAYVMVSPVPARVIGARRDGVSAITLDRGLPADGGDA
Rv0812|M.tuberculosis_H37Rv IYSRGREGGS-APTAYVMVSPVPARVIGARRDGVSAITLDRGLPADGGDA
MMAR_4873|M.marinum_M IYSRGRESGS-APTAYVMVNPVPERVAAVRRDGVAAITLDRGLPAVGADA
MUL_0426|M.ulcerans_Agy99 IYSRGRESGS-APTAYVMVNPVPERVAAVRRAGVAAITLDRGLPAVGADA
MAV_0751|M.avium_104 IYSRGREGGS-VPTAYVMVTAVPDRVTAVRRDGLAAITLDRGLPATGVDA
TH_0072|M.thermoresistible__bu VYSRGRESGAGRPTGYVTVAALPERVAAARRDGVSAITLDRGMASTAAD-
Mflv_1639|M.gilvum_PYR-GCK VYSRGREHGS-EPTAYATIGQVPARVARVRREGLAALTLERGLAADGIDA
Mvan_5108|M.vanbaalenii_PYR-1 VYSRGREHGS-DPTAYATIGSVPARVADVRRNGLAALTLQRSLPAEGVDA
MSMEG_5795|M.smegmatis_MC2_155 VYSRGREGGG-PPTAFATIGALADRVAGARRHGVAAITLDRGLP-VGASE
MAB_0731|M.abscessus_ATCC_1997 VYTRGRESGG-GATAYLTIAPVPERSSRARRDGVSVITLDRGLPAQPAEP
:*:**** *. .*.: : :. * .** *::.:**:*.:. .
Mb0835|M.bovis_AF2122/97 MPWLMASAKTLSYAVNMAVLRHAARQGAGDVIFVSTDGYVLEGPRSTVVI
Rv0812|M.tuberculosis_H37Rv MPWLIASAKTLSYAVNMAVLRHAARQGAGDVIFVSTDGYVLEGPRSTVVI
MMAR_4873|M.marinum_M MPWLLAGAKTLSYAVNMAALRHAANQGAGDVIFVSSDGYVLEGPRSTVVI
MUL_0426|M.ulcerans_Agy99 MPWLLAGAKTLSYAVNMAALRHAANQGAGDVIFVSSDGYVLEGPRSTVVI
MAV_0751|M.avium_104 MPWLLAGAKTLSYAINMAALRHAARHDAGDVVFVSTDGYVLEGPRSTVVI
TH_0072|M.thermoresistible__bu LPWLLAGAKTLSYAVNMAALRHAERHGAGDVIFVSTDGYLLEGPRSTVVI
Mflv_1639|M.gilvum_PYR-GCK MPWLLAGAKTLSYAINMAALRHAERQGAGDVVFVSTDGHILEGPRSTVVI
Mvan_5108|M.vanbaalenii_PYR-1 MPWLLAGAKTLSYAVNMAALRHAERHGAGDVIFISTEGHILEGPRSTVVI
MSMEG_5795|M.smegmatis_MC2_155 MPWLAAGAKTLSYAVNMAALRHAARNGAGDVIFVSSDGFLLEGPRSTVVI
MAB_0731|M.abscessus_ATCC_1997 LPWLLSGAKTLSYAINMSALRYAETQGAQDVIFVSSDGFVLEGPRSTVIV
:*** :.*******:**:.**:* :.* **:*:*::*.:********::
Mb0835|M.bovis_AF2122/97 ATDGDQGGG-NPCLLTPPPWYPILRGTTQQALFEVARAKGYDCDYRALRV
Rv0812|M.tuberculosis_H37Rv ATDGDQGGG-NPCLLTPPPWYPILRGTTQQALFEVARAKGYDCDYRALRV
MMAR_4873|M.marinum_M ATDCDGGAGANPCLLTPPPWYPILRGTTQQALFEVARAKGYDCDYRALRV
MUL_0426|M.ulcerans_Agy99 ATDCDGGAGANPCLLTPPPWYPIVRGTTQQALFEVARAKGYDCDYRALRV
MAV_0751|M.avium_104 ATDSEAPGG-GPCLLTPPPWYPILRGTTQQALFEVARAKGYDCDYRALRV
TH_0072|M.thermoresistible__bu ATEMD---G-VTCLLTPPPWYPILRGTTQQALFEVARNRGYQCDFQPLRP
Mflv_1639|M.gilvum_PYR-GCK ATSSADG---RTCLLTPPPWYPILRGTTQQALFEVARNKGYDCDYQALTP
Mvan_5108|M.vanbaalenii_PYR-1 ATSSPDG---RTCLLTPPPWYPILRGTTQQALFELARNKGYDCDYQALTP
MSMEG_5795|M.smegmatis_MC2_155 ATRGDDG---QPLLLTPPPWYPILRGTTQQALFETARNKGIDCDYRALRP
MAB_0731|M.abscessus_ATCC_1997 DTGDA--------LVTPFPEHGILHGTTQRALFEVAASEGIPCRYEAVKP
* *:** * : *::****:**** * .* * :..:
Mb0835|M.bovis_AF2122/97 ADLFDSQGIWLVSSMTLAARVHTLDGRRLPRTPIAEVFAELVDAAIVSDR
Rv0812|M.tuberculosis_H37Rv ADLFDSQGIWLVSSMTLAARVHTLDGRRLPRTPIAEVFAELVDAAIVSDR
MMAR_4873|M.marinum_M NDLADAHGIWLVSSMTLAARVHTLNGRRLPRSPISEAFSELVDAAVVSDR
MUL_0426|M.ulcerans_Agy99 NDLADAHGIWLVSSMTLAARVHTLNGRRLPRSPISEAFSELVDAAVVSDR
MAV_0751|M.avium_104 ADLFAAQGVWLISSMTLAARVHTLDGRPLPRAPMAAEFAELVDAAIVSDR
TH_0072|M.thermoresistible__bu ADLFAAQGVWLVSSITLGARVHTLDGRPMPGAALAAEMAELVDAAIVSDR
Mflv_1639|M.gilvum_PYR-GCK ADLFAAQGVWLVSSITLAARVHTLDGRPLPQAPLAGDIAALVDAAILCDR
Mvan_5108|M.vanbaalenii_PYR-1 ADLIAAQGVWLVSSITLAARVHTLDGNPLAAAPLSDEIAALVDAAILCDR
MSMEG_5795|M.smegmatis_MC2_155 ADLLTAQGIWLISSITLAARVHTLDGTRLRTSASAPDIAGLVDTAILGDR
MAB_0731|M.abscessus_ATCC_1997 ADLIAAQDVWMLASITLAARVHTLDGVARPAGPLAPRLPELVDKAIAL--
** ::.:*:::*:**.******:* . : :. *** *: