For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPAPTHRVAADVHARIDALLTQVDADLAARFPGDRDAVQPVHTVYIGAADADEHTPRQWGEAALELLERH ADLLTELSDGEGDTALALTRKRLADAPIADLRLDFEDGYGRRSDDVEDADALRAGETLRALGLQSSGVRI KGLTMAERTRAVRTLELVLDGGIPDGFVFTVPKLRAAEQVTAAVWVCEALESAHGLPERTLQFELQIESP QAVLGPDGTATVAAAIHRADGRCTGLHYGTYDYSAACGIAPQHQSLDHPIADHAKAVMQTAAAQTGVWVA DGSTQVSPVGTDEQVAQAIRRHHRLVTRSLERGLYQGWDMHPGHLVTRWLATFAFFRAAMAAAAPRLQNY LDRRGGAIVDEPATAEALATVVLRGLDCGAFEAADVLALAPGATVEVLHDLKNRI
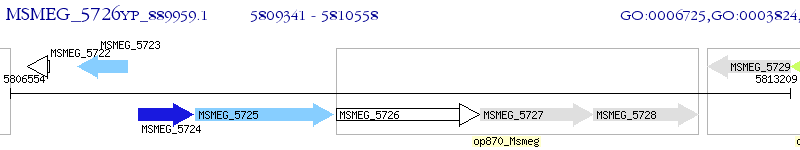
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5726 | - | - | 100% (405) | hypothetical protein MSMEG_5726 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3173 | - | 1e-97 | 49.75% (400) | hypothetical protein Mflv_3173 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2939 | - | 1e-130 | 61.62% (396) | hypothetical protein MAB_2939 |
| M. marinum M | MMAR_1209 | - | 1e-162 | 73.10% (394) | hypothetical protein MMAR_1209 |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2683 | - | 1e-162 | 73.10% (394) | hypothetical protein MUL_2683 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1209|M.marinum_M --------------MAAATENPKSGSRPVDSPA---LAHIDAVLAQVDQE
MUL_2683|M.ulcerans_Agy99 MAVSRVRGICDDGRVAAATENPKSGSRPVDSPA---LAHIDAVLAQVDQE
MSMEG_5726|M.smegmatis_MC2_155 --------------MPAPTHR-------VAADV---HARIDALLTQVDAD
MAB_2939|M.abscessus_ATCC_1997 --------------MAAASSS-----NPVDAGSPGWLTPIAELLTAADAE
Mflv_3173|M.gilvum_PYR-GCK -----------------MTRR-------LDDVV---LADIDRRLAAADAH
: : : * *: .* .
MMAR_1209|M.marinum_M LAARYPGDGPGVQPVHTVYVSAAEATSQTPSQWGGAAAALLQRH--VDLL
MUL_2683|M.ulcerans_Agy99 LAARYPGDGPGVQPVHTVYVSAAEATSQTPSQWGAAAAALLQRH--VDLL
MSMEG_5726|M.smegmatis_MC2_155 LAARFPGDRDAVQPVHTVYIGAADADEHTPRQWGEAALELLERH--ADLL
MAB_2939|M.abscessus_ATCC_1997 LATAYPETRDEPQPIHTVYVSAALADVELPGQWGASALALTARH--EPSL
Mflv_3173|M.gilvum_PYR-GCK LAARYPGDDGRRQPVHTVYIPGNRYSATTPAEWGSTALTAAKDAGGLDAV
**: :* **:****: . * :** :* :
MMAR_1209|M.marinum_M SELGD---------DAALGLVQRRLAAGPIEDLRLDFEDGYGIRGDDVED
MUL_2683|M.ulcerans_Agy99 SELGD---------DAALGLVQRRLAAGPIEDLRLDFEDGYGIRGDDVED
MSMEG_5726|M.smegmatis_MC2_155 TELSDG------EGDTALALTRKRLADAPIADLRLDFEDGYGRRSDDVED
MAB_2939|M.abscessus_ATCC_1997 AALDT---------QGVLPRVKERLAADPIQDLRLDFEDGYGWRGDSTED
Mflv_3173|M.gilvum_PYR-GCK AALVGPNNETDCEPETLATLVEHKLTTEPIEDLRIDFEDGYGTVDDATED
: * : ...:*: ** ***:******* .* .**
MMAR_1209|M.marinum_M DDAVRAGQTLRSLGLPVC-----GVRIKGLTSSEWRRALRTLELVLDG--
MUL_2683|M.ulcerans_Agy99 DDAVRAGQTLRSLGLPVC-----GVRIKGLTSSEWRRALRTLELVLDG--
MSMEG_5726|M.smegmatis_MC2_155 ADALRAGETLRALGLQSS-----GVRIKGLTMAERTRAVRTLELVLD---
MAB_2939|M.abscessus_ATCC_1997 ADARKAGRTLRALSIAANPPAILGIRIKGLTSLHLRRSVRTLELVLDG--
Mflv_3173|M.gilvum_PYR-GCK ADVAQAIATLRSALDAGTSTPFVGTRFKGLEAATRARGLRTLDMFVSGLV
*. :* ***: * *:*** *.:***::.:.
MMAR_1209|M.marinum_M -AGGVPDGFVFTVPKLRAGDQVGAAVQLCEALETAHGLPTGALKFELQIE
MUL_2683|M.ulcerans_Agy99 -AGGVPDGFVFTVPKLRAGDQVGAAVQLCEALETAHGLPTGALKFELQIE
MSMEG_5726|M.smegmatis_MC2_155 --GGIPDGFVFTVPKLRAAEQVTAAVWVCEALESAHGLPERTLQFELQIE
MAB_2939|M.abscessus_ATCC_1997 -ASGVPRGFVTTIPKFRTVTQVEAALRLFDELERVHGLPSGSLRFELQIE
Mflv_3173|M.gilvum_PYR-GCK ASGGLPEGLTLTLPKVTSVDQVEAMVAVASALEDANGLSAGRIRFEVQVE
.*:* *:. *:**. : ** * : : . ** .:**. ::**:*:*
MMAR_1209|M.marinum_M SPQAVIGPDGAATLARAIHMSQRRCTGLHYGTYDYSAACGIAPQHQALDH
MUL_2683|M.ulcerans_Agy99 SPQAVIGPDGAATLARAIHMSQRRCTGLHYGTYDYSAACGIAPQHQALDH
MSMEG_5726|M.smegmatis_MC2_155 SPQAVLGPDGTATVAAAIHRADGRCTGLHYGTYDYSAACGIAPQHQSLDH
MAB_2939|M.abscessus_ATCC_1997 SPQAVLGADGAVTLAKAIHLAQGRLTALHYGTYDYSAACGIAAEQQSLEH
Mflv_3173|M.gilvum_PYR-GCK TPQAILGADGRAPVAQFIHAGQGRVSSLHYGTYDYSASLGIAAAYQSMEH
:***::*.** ..:* ** .: * :.**********: ***. *:::*
MMAR_1209|M.marinum_M PVADHAKAVMLAAAAQTGVRVSDGSTNVVPEGTEEQVVQAFRSHYRLVTR
MUL_2683|M.ulcerans_Agy99 PVADHAKAVMLAAAAQTGVRVSDGSTNVVPEGTEEQVVQAFRSHYRLVTR
MSMEG_5726|M.smegmatis_MC2_155 PIADHAKAVMQTAAAQTGVWVADGSTQVSPVGTDEQVAQAIRRHHRLVTR
MAB_2939|M.abscessus_ATCC_1997 PVADHAKAVMLAAAAQTGVWVCDGSTQVNPEGSPEQQAAALTRHHRLVTR
Mflv_3173|M.gilvum_PYR-GCK PAADHAKNVMQLAVAGTGVHMSDGSTNIVPAGDPDDVAAAWTLHARLVRR
* ***** ** *.* *** :.****:: * * :: . * * *** *
MMAR_1209|M.marinum_M SLERGYYQGWDMHPGHLVTRWLATFTFFRAALGVAAPRLQAYLERAGGGV
MUL_2683|M.ulcerans_Agy99 SLERGYYQGWDMHPGHLVTRWLATFTFFRAALGVAAPRLQAYLERAGGGV
MSMEG_5726|M.smegmatis_MC2_155 SLERGLYQGWDMHPGHLVTRWLATFAFFRAAMAAAAPRLQNYLDRRGGAI
MAB_2939|M.abscessus_ATCC_1997 ALERGYWQGWDMHPGHLVTRWLATYGFHRRALAVAGPRILAYLQRQGGGV
Mflv_3173|M.gilvum_PYR-GCK HLERGIYQGWDMHPAQLVTRYLATYAFYRGAFSPAAARLRNYVHQLDSTV
**** :*******.:****:***: *.* *:. *..*: *:.: .. :
MMAR_1209|M.marinum_M IDEPATAEALAAVVLRGLDCGAFSADQVRDAAPGATESVLRALKMRIPQE
MUL_2683|M.ulcerans_Agy99 IDEPATAEALAAVVLRGLDCGAFSADQVRDAAPGATESVLRALKMRIPQE
MSMEG_5726|M.smegmatis_MC2_155 VDEPATAEALATVVLRGLDCGAFEAADVLALAPGATVEVLHDLKNRI---
MAB_2939|M.abscessus_ATCC_1997 VDEPATAQALATGILRGLDCGAYTDS---ALPPGCDRAVLNQLVQRTHS-
Mflv_3173|M.gilvum_PYR-GCK MDEPATARALAGVIHRGSVCGALTVDEIETALDLPLSTVRDIALGRPTRS
:******.*** : ** *** * *
MMAR_1209|M.marinum_M YNPS
MUL_2683|M.ulcerans_Agy99 YNPS
MSMEG_5726|M.smegmatis_MC2_155 ----
MAB_2939|M.abscessus_ATCC_1997 ----
Mflv_3173|M.gilvum_PYR-GCK TQ--