For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
EGVEWLSAAEYEMWRGYLDSTRLLLRALDRQLEVDAGISFADFEVLALLSEAPERRLRMSEIADAAMTTR SGVTRAVNRLADAGWLRRVECDEDKRGWYAELTEAGFAKVRSSAPVHVAAVRRNVFDLLSPRDIRLFTHA YSEIRAHLLNEADDQP*
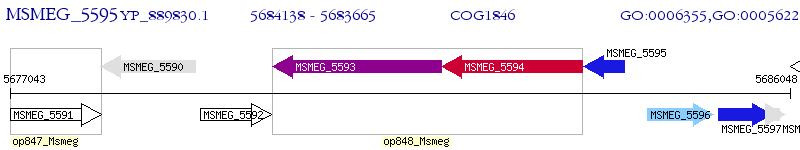
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5595 | - | - | 100% (157) | MarR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_5724 | - | 1e-18 | 35.17% (145) | transcriptional regulator MarR |
| M. smegmatis MC2 155 | MSMEG_6824 | - | 1e-15 | 28.95% (152) | MarR-family protein regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0520 | - | 9e-18 | 36.30% (135) | MarR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0208 | - | 2e-11 | 33.65% (104) | MarR family transcriptional regulator |
| M. marinum M | MMAR_2939 | - | 1e-07 | 32.26% (124) | transcriptional regulatory protein |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1664 | - | 6e-10 | 30.94% (139) | transcriptional regulator, MarR family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0148 | - | 8e-18 | 34.93% (146) | MarR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0520|M.gilvum_PYR-GCK ----------------------------MEWLSDEQQRIWRDYLAMVSKL
Mvan_0148|M.vanbaalenii_PYR-1 ----------------------------MEWLSDEQQLIWRNYLAMVSRL
MSMEG_5595|M.smegmatis_MC2_155 -------------------------MEGVEWLSAAEYEMWRGYLDSTRLL
MAB_0208|M.abscessus_ATCC_1997 --------------------MSTRSESEVPGLDLAEFRSWQNFWVATNRL
TH_1664|M.thermoresistible__bu -------------------------------LD------------AALRL
MMAR_2939|M.marinum_M MSGTRKVPQLPQPIETTRQETLKWVAASAPGLAVDDAELGTALFAGSVRL
* *
Mflv_0520|M.gilvum_PYR-GCK HTAMHRQLQQDCELSLSDYDVLVALSER--GPMRINELGELIGWEQSRLS
Mvan_0148|M.vanbaalenii_PYR-1 HTAMHRQLQQDCELSLSDYDVLVALSER--GPMRINELGELIGWEQSRLS
MSMEG_5595|M.smegmatis_MC2_155 LRALDRQLEVDAGISFADFEVLALLSEAPERRLRMSEIADAAMTTRSGVT
MAB_0208|M.abscessus_ATCC_1997 NFLLNRKMVASHSISLTDFHVLQQLLRSANGSCRMGDIANSLLASRSRIT
TH_1664|M.thermoresistible__bu YATLNKTLVDEHHLTLNDVRLLDMLDKSPSGSARMGDLADALMSLPSRVT
MMAR_2939|M.marinum_M VRTVESHLQR-YGLSSGRFAVLITLGAAPEGRHTPSAIAERIAVRRPTVT
:. : :: :* * . :.: . ::
Mflv_0520|M.gilvum_PYR-GCK HQLRRMRGRGLVEREGDGDDRRGATVALTDAGLAAVQTAAPGHVDLVRTV
Mvan_0148|M.vanbaalenii_PYR-1 HQLRRMRGRGLVGREGDGDDRRGATVALTDAGLAALRTAAPGHVDLVRTM
MSMEG_5595|M.smegmatis_MC2_155 RAVNRLADAGWLRRVECDEDKRGWYAELTEAGFAKVRSSAPVHVAAVRRN
MAB_0208|M.abscessus_ATCC_1997 HQVRRLEEQGLVKRGSMPQDRRAVLAALTEQGRTLGEAAMRTYSECVREH
TH_1664|M.thermoresistible__bu RQIRRLEAHNLVRRCASPDDGRGVLAIITDAGREEVRKAMVTYGNEVRVH
MMAR_2939|M.marinum_M GIVNGLESAGLVRRSADPTSRRNQLVELTERGRRLVAEIAPDHFGRLAAA
:. : . : * . * . :*: * : :
Mflv_0520|M.gilvum_PYR-GCK V--FDGLSKAEERAFGATIAAVLERLNARSAG--------
Mvan_0148|M.vanbaalenii_PYR-1 V--FDGLTPAAQREFGAAVETVLSRLRAAGSTD-------
MSMEG_5595|M.smegmatis_MC2_155 V--FDLLSPRDIRLFTHAYSEIRAHLLNEADDQP------
MAB_0208|M.abscessus_ATCC_1997 Y--LMRLTRPQQAAIAESFRRVGEGAEEALHDGDGRGGE-
TH_1664|M.thermoresistible__bu F--LSRLSRPQIAAMGENCRRISAGLRADGP--PARLGRV
MMAR_2939|M.marinum_M MGEFTPAERDTLRAAMGLLDRFGEVLLDERDA--------
: .