For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNRAVRHTGLMRSIWKGSIAFGLVNVPVKVYSATEDHDIKFHQVHAKDNGRIRYKRVCEVCGEVVEYRDI NKAFESDDGQMVVITDEDIATLPEERSREIEVVEFIPAEQLDPLMYDKSYFLEPDSKSSKSYVLLAKTLA ETDRIAIVHFSLRNKSRLAALRVKDFSKRDVMMIHTLLWPDEIRDPDFPILDKEVQIKPAELKMAGQVVE SMTDDFKPDLYHDDYQEQLRELVQAKLEGGEAFSVEEQPAELDEGTEDVSDLLAKLEASVKARKGGKSDS KDDSDSESDSKESKSDSKPAKKAPAKKAAAKKSTAKKAPAKKAAAKKS
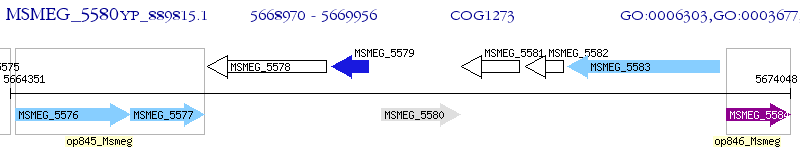
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5580 | - | - | 100% (328) | Ku protein |
| M. smegmatis MC2 155 | MSMEG_6438 | - | 1e-08 | 56.90% (58) | hypothetical protein MSMEG_6438 |
| M. smegmatis MC2 155 | MSMEG_2389 | hup | 2e-05 | 75.76% (33) | DNA-binding protein HU |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0962c | - | 1e-126 | 79.93% (269) | hypothetical protein Mb0962c |
| M. gilvum PYR-GCK | Mflv_1826 | - | 1e-149 | 81.40% (328) | Ku domain-containing protein |
| M. tuberculosis H37Rv | Rv0937c | - | 1e-126 | 79.93% (269) | hypothetical protein Rv0937c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1032c | - | 1e-122 | 78.52% (270) | hypothetical protein MAB_1032c |
| M. marinum M | MMAR_4575 | - | 1e-136 | 79.60% (299) | hypothetical protein MMAR_4575 |
| M. avium 104 | MAV_1050 | - | 1e-142 | 80.06% (316) | Ku protein |
| M. thermoresistible (build 8) | TH_0600 | - | 1e-147 | 83.92% (311) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4436 | - | 1e-134 | 78.60% (299) | hypothetical protein MUL_4436 |
| M. vanbaalenii PYR-1 | Mvan_4917 | - | 1e-143 | 80.19% (318) | Ku domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0962c|M.bovis_AF2122/97 ----------MRAIWTGSIAFGLVNVPVKVYSATADHDIRFHQVHAKDNG
Rv0937c|M.tuberculosis_H37Rv ----------MRAIWTGSIAFGLVNVPVKVYSATADHDIRFHQVHAKDNG
MMAR_4575|M.marinum_M ----------MRSIWKGSIAFGLVNVPVKVYSATQDHDIKFHQVHAKDNG
MUL_4436|M.ulcerans_Agy99 ----------MRSIWKGSIAFGLVNVPVKVYSATQDHDIKFHQVHAKDNG
MAV_1050|M.avium_104 ----------MRSIWKGSIAFGLVNVPVKVYSATEDHDIKFHQVHAKDNG
Mflv_1826|M.gilvum_PYR-GCK ----------MRSIWKGSIAFGLVNVPVKVYSATEDHDIKFHQVHEKDNG
Mvan_4917|M.vanbaalenii_PYR-1 ----------MRSIWKGSIAFGLVNVPVKVYSATEDHDIKFHQVHDKDNG
TH_0600|M.thermoresistible__bu ----------MRSIWKGSIAFGLVNVPVKVYSATQDHDVKFHQVHAKDNG
MSMEG_5580|M.smegmatis_MC2_155 MNRAVRHTGLMRSIWKGSIAFGLVNVPVKVYSATEDHDIKFHQVHAKDNG
MAB_1032c|M.abscessus_ATCC_199 ----------MRSIWKGSLAFGLVNVPVKVYSATEDHDLKFHQVHNKDNG
**:**.**:*************** ***::***** ****
Mb0962c|M.bovis_AF2122/97 RIRYKRVCEACGEVVDYRDLARAYESGDGQMVAITDDDIASLPEERSREI
Rv0937c|M.tuberculosis_H37Rv RIRYKRVCEACGEVVDYRDLARAYESGDGQMVAITDDDIASLPEERSREI
MMAR_4575|M.marinum_M RIRYKRVCEVCGEVVEYSDLARAFESDDGQMVIINDDDLATLPEDRSREI
MUL_4436|M.ulcerans_Agy99 RIRYKRVCEVCGEVVEYSDLARAFESDDGQMVIINDDDLATLPEDRSREI
MAV_1050|M.avium_104 RIRYQRVCELDGEVVEYRDIARAYESDDGQMVIITDDDIATLPEERSREI
Mflv_1826|M.gilvum_PYR-GCK RIRYNRVCEVCGEVVEYRDIAKAYESDDGQTVIITDDDISTLPEERSREI
Mvan_4917|M.vanbaalenii_PYR-1 RIRYKRVCEVCGEVVEYRDIAKAYESDDGQTVIITDDDISTLPEERSREI
TH_0600|M.thermoresistible__bu RIRYKRVCEVCGEVVEYRDIARAYESDDGQMVVITDEDLATLPEERSREI
MSMEG_5580|M.smegmatis_MC2_155 RIRYKRVCEVCGEVVEYRDINKAFESDDGQMVVITDEDIATLPEERSREI
MAB_1032c|M.abscessus_ATCC_199 RIRYKRTCEECGEVVEFRDINKAYEAEDGRTVVITDEDLASLPAESSREI
****:*.** ****:: *: :*:*: **: * *.*:*:::** : ****
Mb0962c|M.bovis_AF2122/97 EVLEFVPAADVDPMMFDRSYFLEPDSKSSKSYVLLAKTLAETDRMAIVHF
Rv0937c|M.tuberculosis_H37Rv EVLEFVPAADVDPMMFDRSYFLEPDSKSSKSYVLLAKTLAETDRMAIVHF
MMAR_4575|M.marinum_M EVLEFVPAEEVDPMMIDRSYFLEPDSKSSKSYVLLAKTLAETERMAIVHF
MUL_4436|M.ulcerans_Agy99 EVLEFVPAEEVDPMMIDRSYFLEPDSESSKSYVLLAKTLAETERMAIVHF
MAV_1050|M.avium_104 EVLEFVPANEVDPMLFDRSYFLEPDSKSSKSYVLLAKTLAETDRMAIVHF
Mflv_1826|M.gilvum_PYR-GCK EVLEFVPAADLDPLMYDRSYFLEPEGKSTKSYVLLAKTLAETDRVAIVNF
Mvan_4917|M.vanbaalenii_PYR-1 EVLEFVPAGDLDPLMYDRSYFLEPDGKSSKSYVLLAKTLMDTDRVAIVNF
TH_0600|M.thermoresistible__bu EVVEFVPAEQLDPMMYDRSYFLEPDSKSSKSYVLLAKTLAQTDRIAIVHF
MSMEG_5580|M.smegmatis_MC2_155 EVVEFIPAEQLDPLMYDKSYFLEPDSKSSKSYVLLAKTLAETDRIAIVHF
MAB_1032c|M.abscessus_ATCC_199 EVLEFIPAAELDPLMYDKSYYLEPDSKSSKSYVLLAQTLAQTDRVAIVHF
**:**:** ::**:: *:**:***:.:*:*******:** :*:*:***:*
Mb0962c|M.bovis_AF2122/97 TLRNKTRLAALRVKDFGKREVMMVHTLLWPDEIRDPDFPVLDQKVEIKPA
Rv0937c|M.tuberculosis_H37Rv TLRNKTRLAALRVKDFGKREVMMVHTLLWPDEIRDPDFPVLDQKVEIKPA
MMAR_4575|M.marinum_M TLRSKTRLAALRVKDFGKREVMVVHTLLWPDEIRDPDFPVLDKEVEIKPA
MUL_4436|M.ulcerans_Agy99 TLRSKTRLAALRAKDFGKREVMVVHTLLWLDEIRDPDFPVLDKEVEIKPA
MAV_1050|M.avium_104 TLRNKTRLAALRVKDFGKRDVMVIHTLLWPDEIRDPDFPILDKKVEIKPA
Mflv_1826|M.gilvum_PYR-GCK ALRNKTRLAALRVKDFSKRDVMVIHTLLWPDEIRDPDFPVLDKDVDVKPA
Mvan_4917|M.vanbaalenii_PYR-1 ALRNKTRLAALRVKDFSKRDVMMIHTLLWPDEIRDPDFPVLDKKVDVKPA
TH_0600|M.thermoresistible__bu SLRNKTRLGALRVKDFSKRDVMMIHTLLWPDEIRDPDFPVLDKEVEIKPA
MSMEG_5580|M.smegmatis_MC2_155 SLRNKSRLAALRVKDFSKRDVMMIHTLLWPDEIRDPDFPILDKEVQIKPA
MAB_1032c|M.abscessus_ATCC_199 ALRNKTRLAALRVKDFGERQVMIIHTLLWPDEIREPDFPVLDQEVKIKPA
:**.*:**.***.***.:*:**::***** ****:****:**:.*.:***
Mb0962c|M.bovis_AF2122/97 ELKMAGQVVDSMADDFNPDRYHDTYQEQLQELIDTKLEGGQAFTAEDQPR
Rv0937c|M.tuberculosis_H37Rv ELKMAGQVVDSMADDFNPDRYHDTYQEQLQELIDTKLEGGQAFTAEDQPR
MMAR_4575|M.marinum_M ELKMAGQVVESMADDFNPDRYRDTYQEQLQELIDAKLEGGEAFTAEEQPA
MUL_4436|M.ulcerans_Agy99 ELKMAGQVVESMADDFNPDRYRDTYQEQLQELIDAKLEGGEAFTAEEQPA
MAV_1050|M.avium_104 ELKMAGQVVESMAEDFNPDRYHDDYQEQLHELVQAKLEGGEAFTTEEQPK
Mflv_1826|M.gilvum_PYR-GCK ELKMASQVVDSMTDDFNPDRYHDDYQEQLRELVQAKLEGGEAFTTEEQPQ
Mvan_4917|M.vanbaalenii_PYR-1 ELKMASQVVDSMTDDFNPDRYHDDYQEQLHELIQAKLEGGEAFTTEEQPQ
TH_0600|M.thermoresistible__bu ELKMAAQVVDSMTDDFHPDQYRDTYQEQLQELVQAKLEGGEAFTTEEQPT
MSMEG_5580|M.smegmatis_MC2_155 ELKMAGQVVESMTDDFKPDLYHDDYQEQLRELVQAKLEGGEAFSVEEQPA
MAB_1032c|M.abscessus_ATCC_199 ELAMAGQVVESMADDFDPSRYNDTYQSQLLEMVEAKLEGGEAFTKEEAPT
** **.***:**::**.*. *.* **.** *::::*****:**: *: *
Mb0962c|M.bovis_AF2122/97 LLDE-PEDVSDLLAKLEASVKARS--------------------------
Rv0937c|M.tuberculosis_H37Rv LLDE-PEDVSDLLAKLEASVKARS--------------------------
MMAR_4575|M.marinum_M ELDE-TEDVSDLLAKLEASVKARSG-------------------------
MUL_4436|M.ulcerans_Agy99 ELDE-TEDVSDLLAKLEASVKARSG-------------------------
MAV_1050|M.avium_104 QLDE-TEDVSDLLAKLEASVKARSG-------------------------
Mflv_1826|M.gilvum_PYR-GCK ELDE-SEDVSDLLAKLEASVKARKAQSSGKSSDSDDSDDSDDSDDSDDSD
Mvan_4917|M.vanbaalenii_PYR-1 ELDE-TEDVSDLLAKLEASVKARKAQGT----------------------
TH_0600|M.thermoresistible__bu ELDE-TEDVSDLLAKLEASVKARRSGGSSKSS------------------
MSMEG_5580|M.smegmatis_MC2_155 ELDEGTEDVSDLLAKLEASVKARKGGKSDSKDDSDS------------ES
MAB_1032c|M.abscessus_ATCC_199 ELDA--TDVSDLLAKLEASVKKRQ--------------------------
** ************** *
Mb0962c|M.bovis_AF2122/97 ---KANSNVPTPP---------------------------
Rv0937c|M.tuberculosis_H37Rv ---KANSNVPTPP---------------------------
MMAR_4575|M.marinum_M -DGKSAAKKSTAEKAPAKKARAKQAAKS------------
MUL_4436|M.ulcerans_Agy99 -DGKSAAKKSTAERAPAKKARAKQAAKS------------
MAV_1050|M.avium_104 -DGKAPARKSPAKKTAAKKAPAKKGAAKKAPAKKAASRS-
Mflv_1826|M.gilvum_PYR-GCK EKDDKPAKKVPAKKSAAKKAPAKK-AAKKAPAKKAAAKKG
Mvan_4917|M.vanbaalenii_PYR-1 QKDEPAAEKPAAKKPAAKKSPAKKTAAKKAPAKKAPAKK-
TH_0600|M.thermoresistible__bu DADDGDKPKKSPAKKAAKKAPAKKAARKASAKKTAPKK--
MSMEG_5580|M.smegmatis_MC2_155 DSKESKSDSKPAKKAPAKKAAAKKSTAKKAPAKKAAAKKS
MAB_1032c|M.abscessus_ATCC_199 ------AEKASTN---------------------------
..