For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSAVVQVRGLRRSFGAQQVLDDLELQIDDGEFVAMLGRSGSGKSTLLRILAGLDDDVTGSVVVPRSRAVV FQNPRLLPWRRALANVTFALRDGGPDEPSRTARARAALDEVGLSDKARSWPLSLSGGEAQRVSLARALVR EPDLLLLDEPFGALDALTRLKMYRLLHELWARRHMAVLHVTHDVDEAILLADRVVVLSGGQVSLDRRVDL PFPRSRGDDGFDDLRRVLLAELGVREEVGHDR
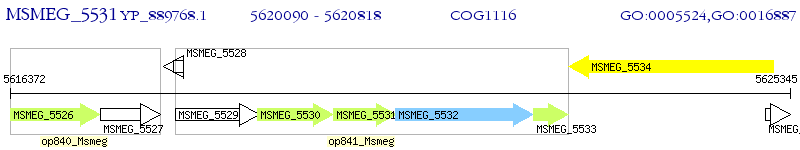
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5531 | - | - | 100% (242) | ABC transporter ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_4587 | - | 2e-58 | 52.81% (231) | putative aliphatic sulfonates transport ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_3853 | - | 2e-56 | 51.90% (237) | putative aliphatic sulfonates transport ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2856c | ugpC | 4e-31 | 39.41% (203) | sn-glycerol-3-phosphate transport ATP-binding protein ABC |
| M. gilvum PYR-GCK | Mflv_3158 | - | 1e-116 | 88.38% (241) | ABC transporter related |
| M. tuberculosis H37Rv | Rv2832c | ugpC | 4e-31 | 39.41% (203) | sn-glycerol-3-phosphate transport ATP-binding protein ABC |
| M. leprae Br4923 | MLBr_00669 | ftsE | 2e-27 | 40.78% (206) | putative cell division ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_2217 | - | 1e-60 | 53.85% (234) | sulfonate ABC transporter, ATP-binding subunit SsuB |
| M. marinum M | MMAR_3715 | cysA1 | 1e-29 | 37.32% (209) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. avium 104 | MAV_2434 | - | 1e-59 | 54.08% (233) | putative aliphatic sulfonates transport ATP-binding protein |
| M. thermoresistible (build 8) | TH_3596 | - | 5e-61 | 57.14% (238) | PUTATIVE ABC transporter related |
| M. ulcerans Agy99 | MUL_3658 | cysA1 | 1e-29 | 37.32% (209) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. vanbaalenii PYR-1 | Mvan_3412 | - | 2e-60 | 53.62% (235) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5531|M.smegmatis_MC2_155 ---------------MSAVVQVRGLRRSFG--AQQVLDDLELQIDDGEFV
Mflv_3158|M.gilvum_PYR-GCK --------------MSSPVVTVRGLHRAFG--AQQVLDDLDLDIAGGEFV
TH_3596|M.thermoresistible__bu ---MAAGPGRLTGNRAEPVAVVRGLHRSFG--AGAVLDGIDLTIRRGEFV
MAV_2434|M.avium_104 MTLTAESGP-LAGSRTDVAGELRHVDKWYG--NRHVLQDVSLQIPSGQIV
Mvan_3412|M.vanbaalenii_PYR-1 MTTTSHAAERGTAAHTTAAGELRNVNKWYG--VHHVLTDVSVRVACGEIV
MAB_2217|M.abscessus_ATCC_1997 MTAVFEVTE-LSPHSTTAAAELTSVTKWYG--RNRVLDDVSLRVGCGEIV
MMAR_3715|M.marinum_M ---MKKETSLTGPGTGGHAIVVRDAFKQYG--DFVALDHVDFVVPAGSLT
MUL_3658|M.ulcerans_Agy99 ---------MTGPGTGGHAIVVRDAFKQYG--DFVALDHVDFVVPAGSLT
MLBr_00669|M.leprae_Br4923 ------------------MITLDHVSKKYKSLARPALDNVNVKIDKGEFV
Mb2856c|M.bovis_AF2122/97 ----------------MANVQYSAVTQRYPGADAPTVDNLDLDIADGEFL
Rv2832c|M.tuberculosis_H37Rv ----------------MANVQYSAVTQRYPGADAPTVDNLDLDIADGEFL
: : .: :.. : *.:
MSMEG_5531|M.smegmatis_MC2_155 AMLGRSGSGKSTLLRILAGLDDDVTG-----SVVVPRSR-----------
Mflv_3158|M.gilvum_PYR-GCK AMLGRSGSGKSTLLRILAGLDNRADG-----SVLVPRSR-----------
TH_3596|M.thermoresistible__bu ALLGRSGSGKSTLLRALAGLDHGGAGGGGG-DVYVSRDV-----------
MAV_2434|M.avium_104 ALIGRSGSGKSTVLRVLAGLSHDHTG-----RRLVAGAP-----------
Mvan_3412|M.vanbaalenii_PYR-1 ALIGRSGSGKSTVLRVLAGLSTDHTG-----DRAVAGAP-----------
MAB_2217|M.abscessus_ATCC_1997 ALVGRSGSGKSTVLRVLSGLAGDHTG-----ERKVLGAP-----------
MMAR_3715|M.marinum_M ALLGPSGSGKSTLLRTIAGLDQPDSGNITINGRDVTRVPP-----QRRGI
MUL_3658|M.ulcerans_Agy99 ALLGPSGSGKSTLLRTIAGLDQPDSGNITINGRDVTRVPP-----QRRGI
MLBr_00669|M.leprae_Br4923 FLIGPSGSGKSTFMRLLLGAETPTSGDVRVSKFHVNKLPGRHIPRLRQVI
Mb2856c|M.bovis_AF2122/97 VLVGPSGCGKSTTLRVLAGLEPIESGRISIGDVDVTHLPP-----RARDV
Rv2832c|M.tuberculosis_H37Rv VLVGPSGCGKSTTLRVLAGLEPIESGRISIGDVDVTHLPP-----RARDV
::* **.**** :* : * * *
MSMEG_5531|M.smegmatis_MC2_155 AVVFQNPRLLPWRRALANVTFALRDGGPDEPSRTARARAALDEVGLSDKA
Mflv_3158|M.gilvum_PYR-GCK AVVFQNPRLLPWRRALANVTFALPQAGPDAPSRTARGQAALDEVGLSGKA
TH_3596|M.thermoresistible__bu AVVFQDSRLLPWARVLDNVTLGLS--GRDAER---RGAAVLADVGLAGRE
MAV_2434|M.avium_104 ALAFQEPRLFPWRDVRTNVGYGLTRTRLPRAQVRRRAERALADVGLADHA
Mvan_3412|M.vanbaalenii_PYR-1 ALAFQEPRLFPWRDVRTNVVYGLTRARLTRAEAVARADRALADVGLSDRA
MAB_2217|M.abscessus_ATCC_1997 AVAFQEPRLFPWRSVRDNVLYGLTRTKLSRDEALSRVDRALDEVGLADKA
MMAR_3715|M.marinum_M GFVFQHYAAFKHLTVRDNVAYGLKIRKRPKAEIRDKVDNLLEVVGLSGFH
MUL_3658|M.ulcerans_Agy99 GFVFQHYAAFKHLTVRDNVAYGLKIRKRPKAEIRDKVDNLLEVVGLSGFH
MLBr_00669|M.leprae_Br4923 GCVFQDFRLLQQKTVYENVAFALEVIGRRSDAINQVVPDVLETVGLSGKA
Mb2856c|M.bovis_AF2122/97 AMVFQNYALYPNMTVAANMGFALRNAGMSRADTRRRVLEVADMLELTDLL
Rv2832c|M.tuberculosis_H37Rv AMVFQNYALYPNMTVAANMGFALRNAGMSRADTRRRVLEVADMLELTDLL
. .**. . *: .* : *:.
MSMEG_5531|M.smegmatis_MC2_155 RSWPLSLSGGEAQRVSLARALVREPDLLLLDEPFGALDALTRLKMYRLLH
Mflv_3158|M.gilvum_PYR-GCK RAWPLSLSGGEAQRVSLARALVREPDLLLLDEPFGALDALTRLKMYRLLH
TH_3596|M.thermoresistible__bu RAWPYELSGGEQQRVALARSLVRDPALLLADEPFGALDALTRIRMHRLLQ
MAV_2434|M.avium_104 RAWPLTLSGGQAQRVSLARALVAEPRLLLLDEPFGALDALTRLSMHTLLL
Mvan_3412|M.vanbaalenii_PYR-1 AAWPLTLSGGQAQRVSLARALVAEPTLLLLDEPFGALDALTRLSMRTLLL
MAB_2217|M.abscessus_ATCC_1997 QAWPLTLSGGQAQRVSLARALVAEPQLLLLDEPFGALDALTRLSMHRLLL
MMAR_3715|M.marinum_M GRYPNQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRAWLR
MUL_3658|M.ulcerans_Agy99 GRYPDQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRAWLR
MLBr_00669|M.leprae_Br4923 NRLPDELSGGEQQRVAIARAFVNRPLVLLADEPTGNLDPDTSKDIMDLLE
Mb2856c|M.bovis_AF2122/97 DRKPAKLSGGQRQRVAMGRAIVRRPRVFCMDEPLSNLDAKLRVSTRSQIS
Rv2832c|M.tuberculosis_H37Rv DRKPAKLSGGQRQRVAMGRAIVRRPRVFCMDEPLSNLDAKLRVSTRSQIS
* ****: **:::.*::. * :: *** . **. :
MSMEG_5531|M.smegmatis_MC2_155 ELWARRHMAVLHVTHDVDEAILLADRVVVLSGGQVSLDRRVDLPFPRSRG
Mflv_3158|M.gilvum_PYR-GCK DLWARRRMAVLHVTHDVDEAILLADRVVVLSDGAVSLDRRVDLPFPRSRG
TH_3596|M.thermoresistible__bu ELCAKYRPGVLLVTHDVDEAVTLADRVLVLADGRFVTDRELTFRGRRTLR
MAV_2434|M.avium_104 DLWRRHGFGVLLVTHDVDEAVALADRVLVLEDGRVVHELAIDPP-RRTPG
Mvan_3412|M.vanbaalenii_PYR-1 DLWREHGFGVLLVTHDVDEAVALADRVVVLEDGAVVHTLDIPDP-RRATG
MAB_2217|M.abscessus_ATCC_1997 NLWEQHGFGVLLVTHDVDEAISLADTVLVLDEGRLAHTLRIRNP-RKPRG
MMAR_3715|M.marinum_M RLHDEVHVTTVLVTHDQAEALDVADRIAVLNSGRIEQVGSPTEVYDAPAN
MUL_3658|M.ulcerans_Agy99 RLHDEVHVTTVLVTHDQAEALDVADRIAVLNSGRIEQVGSPTEVYDAPTN
MLBr_00669|M.leprae_Br4923 RIN-RTGTTVLMATHDHHIVDSMRQRVVELSLGRLVRDEWCGIYGMDR--
Mb2856c|M.bovis_AF2122/97 GLQRRLGTTTVYVTHDQVEAMTMGDRVAVLKDGVLQQVDTPRALYDDPVN
Rv2832c|M.tuberculosis_H37Rv GLQRRLGTTTVYVTHDQVEAMTMGDRVAVLKDGVLQQVDTPRALYDDPVN
: . .: .*** . : : : * * .
MSMEG_5531|M.smegmatis_MC2_155 DDG--FDDLRRVLLAELGVREEVGHDR-----------------------
Mflv_3158|M.gilvum_PYR-GCK DDG--FDELRRELLAELGVKEEVGHDRSR---------------------
TH_3596|M.thermoresistible__bu NPE--FVTHREALLAALGVGDD----------------------------
MAV_2434|M.avium_104 EPGAHTERYRAELLDRLGVRQ-----------------------------
Mvan_3412|M.vanbaalenii_PYR-1 EG--YTDRYRAELLDKLGVQI-----------------------------
MAB_2217|M.abscessus_ATCC_1997 KHDAETENHRGELLAQLGV-------------------------------
MMAR_3715|M.marinum_M AFVMSFLGAVSALNGALVRPHDIRVGRTPEMAVAAADGTGESTGVTRAVV
MUL_3658|M.ulcerans_Agy99 AFVMSFLGAVSALNGALVRPHDIRVGRTPEMAVAAADGTGESTGVTRAVV
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb2856c|M.bovis_AF2122/97 TFVATFIGAPAMNLIDAAVAHGVVRAPDLAIPVPDPAAERVLVGVRPESW
Rv2832c|M.tuberculosis_H37Rv TFVATFIGAPAMNLIDAAVAHGVVRAPDLAIPVPDPAAERVLVGVRPESW
MSMEG_5531|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3158|M.gilvum_PYR-GCK --------------------------------------------------
TH_3596|M.thermoresistible__bu --------------------------------------------------
MAV_2434|M.avium_104 --------------------------------------------------
Mvan_3412|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2217|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3715|M.marinum_M DRIVVLGFEVRVELTSAATGGAFTAQITRGDAEALALQEGDTVYVRATRV
MUL_3658|M.ulcerans_Agy99 DRIVVLGFEVRVELTSAATGGAFTAQITRGDAEALALQEGDTVYVRATRV
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mb2856c|M.bovis_AF2122/97 DVASIGTPGSLTVHVELVEELGFESFVYATPVDQRGWSSRAPRIVFRTDR
Rv2832c|M.tuberculosis_H37Rv DVASIGTPGSLTVHVELVEELGFESFVYATPVDQRGWSSRAPRIVFRTDR
MSMEG_5531|M.smegmatis_MC2_155 -------------------------------
Mflv_3158|M.gilvum_PYR-GCK -------------------------------
TH_3596|M.thermoresistible__bu -------------------------------
MAV_2434|M.avium_104 -------------------------------
Mvan_3412|M.vanbaalenii_PYR-1 -------------------------------
MAB_2217|M.abscessus_ATCC_1997 -------------------------------
MMAR_3715|M.marinum_M PPIAGAAPAVPDDGAGSTPSAAQIGVGGQ--
MUL_3658|M.ulcerans_Agy99 PPIAGAAPAVPDDGAGSTPRAAQIGVGGQ--
MLBr_00669|M.leprae_Br4923 -------------------------------
Mb2856c|M.bovis_AF2122/97 RTAVRVGESLAIVPHSQEVRLFNSRTETRLR
Rv2832c|M.tuberculosis_H37Rv RTAVRVGESLAIVPHSQEVRLFNSRTETRLR