For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLGELVPGPRERVADAVGATEARRTHRGVTPGLTEAACGRVSRAWSARVTRRRVARRSRRRRRRIASALR RRRRWVTAGGRRREATLRRRRVASRRRGWRVTALRWRAETARLRRGIAAVRRRGRRVAAVGRRGRRVAAV GRRRVVARRLISHVASSPHLG
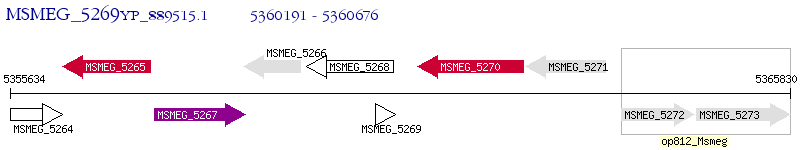
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5269 | - | - | 100% (161) | hypothetical protein MSMEG_5269 |
| M. smegmatis MC2 155 | MSMEG_2627 | - | 2e-05 | 37.69% (130) | hypothetical protein MSMEG_2627 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1363 | - | 2e-05 | 34.48% (145) | LigA |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5269|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1363|M.vanbaalenii_PYR-1 MSRGSNHGNSSILVGRRRCGHRECCGGSPGDRTHHVHPGRGRRVCDGDPA
MSMEG_5269|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1363|M.vanbaalenii_PYR-1 QRGTGRQFAVVGVGTARHRRADGGSAADQCGLRRVRRSRSGRVRRCADLR
MSMEG_5269|M.smegmatis_MC2_155 ---------------------------------------------MLGEL
Mvan_1363|M.vanbaalenii_PYR-1 PNRTESCCRRRIRVPRRTGGGVPRIGAGRGAGLSGRGGERLGVRRRLRAV
* :
MSMEG_5269|M.smegmatis_MC2_155 VPGP---RERVADAVGATEARRTHRGVTPGLTEAACG-RVSRAWSARVTR
Mvan_1363|M.vanbaalenii_PYR-1 RSGRSHLRRRPAGCGGVCLAARQRRGVRRGFRPVATDRRVLRRRPARCGG
.* *.* *.. *. * * :*** *: .* . ** * .**
MSMEG_5269|M.smegmatis_MC2_155 RRVARRSRRRRRR----IASALRRRRRWVTAGGRRREATLRRR-------
Mvan_1363|M.vanbaalenii_PYR-1 VRVAACQRRGFRRGLLPVGGDRRVLRRWTRCGGRDRLTGAQRCGVRRELV
*** .** ** :.. * ***. .*** * : :*
MSMEG_5269|M.smegmatis_MC2_155 -----RVASRRRGWRVTALRWRAETARLR---------------------
Mvan_1363|M.vanbaalenii_PYR-1 PLGCDRVVLRRLSRRRHHRHLRRHPVRVRRHHRNGRPGRRAGRRGGRGGG
**. ** . * : * ...*:*
MSMEG_5269|M.smegmatis_MC2_155 -----------------------------------------RGIAAVRRR
Mvan_1363|M.vanbaalenii_PYR-1 RRGPGGAGGGGAGRHGGSDGARSGGCGRHVRPRGGRRDRAGRGFPGLRRR
**:..:***
MSMEG_5269|M.smegmatis_MC2_155 GRRVAA--------------------------------VGRRGRRVAAVG
Mvan_1363|M.vanbaalenii_PYR-1 GGRGAGGDHGAGVERGHGTGGGFGAGAGRDHGGGGGFRDGRRGRDHGGGG
* * *. ***** .. *
MSMEG_5269|M.smegmatis_MC2_155 RRR-----------------VVARRLISHVASSPHLG-------------
Mvan_1363|M.vanbaalenii_PYR-1 GFRDGRRGRDHGGGGGFGVGVGRGRNLRHSGEVRHSGEVRHGGEVGQCRH
* * * : * .. * *
MSMEG_5269|M.smegmatis_MC2_155 -------------------
Mvan_1363|M.vanbaalenii_PYR-1 LGQGRSGVGERRPALTARP