For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TFTRLLPCRIDDILLRELDYRDAEAFALGTTDPAVQEYGHLPLTHYTPEIVRDQIDGIIAQGLADGSLAV LAIADADSDAFLGSLVLFDFRSDRAEVGFWLTAQSRGRGAAVKALAASSQIVATMGLELLTARTSPQNLA SQRVLERAGFQQKGMPTEQQTPSGVRVPVLTFERPVVEFRFNV*
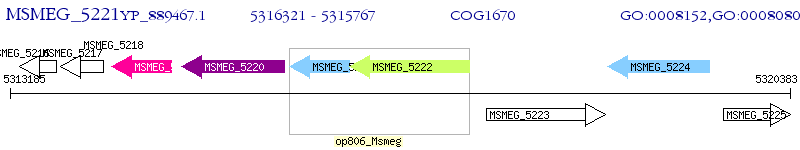
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5221 | - | - | 100% (184) | GCN5-related N-acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00184 | rimJ | 5e-05 | 32.26% (62) | putative acetyltransferase |
| M. abscessus ATCC 19977 | MAB_4621c | - | 8e-06 | 28.67% (150) | putative acetyltransferase |
| M. marinum M | MMAR_4519 | rimJ | 5e-05 | 32.00% (75) | ribosomal-protein-alanine acetyltransferase RimJ |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1242 | - | 2e-56 | 65.68% (169) | GCN5-related N-acetyltransferase |
| M. ulcerans Agy99 | MUL_4691 | rimJ | 3e-05 | 32.00% (75) | ribosomal-protein-alanine acetyltransferase RimJ |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4519|M.marinum_M MNLMRAGQRHPGWPMDVGPLRVRAGVIRLRPVRMRDGAQWSRTRLADRAH
MUL_4691|M.ulcerans_Agy99 --------------MDVGPLRVRAGVIRLRPVRMRDGAQWSRTRLADRAH
MLBr_00184|M.leprae_Br4923 ---MRFNARHPGWPSNVGPLRVPAGVIRLRAVRLRDGVQWSRIRLADRAY
MSMEG_5221|M.smegmatis_MC2_155 -------------MTFTRLLPCRIDDILLRELDYRDAEAFALG--TTDPA
TH_1242|M.thermoresistible__bu --------------VIAEHLPCRAGDVVVRRLDHADAEAFAAG--TTDTA
MAB_4621c|M.abscessus_ATCC_199 ----------------MEPVELQTDRLMLRAVADADAAAIVDA--CNDPE
: . : :* : *. .
MMAR_4519|M.marinum_M LEPWEPSAEGDWTIRHSVAAWPAVCSGLRSEARKGRMLPYVIELDGRFCG
MUL_4691|M.ulcerans_Agy99 LEPWEPSAEGDWTIRHSVAAWPAVCSGLRSEARKGRMLPYVIELDGRFCG
MLBr_00184|M.leprae_Br4923 LEPWEPSTEGDWVVRHSVIAWQVLCSSLRSEARKGRMLPYAIELDGNFCG
MSMEG_5221|M.smegmatis_MC2_155 VQEYGHLPLTHYTPEIVRDQIDGIIAQGLADG-SLAVLAIADADSDAFLG
TH_1242|M.thermoresistible__bu VRQYGHLPLREYTPQTVRDQIDGVIADGLADG-SLAVLAIADASSNQFLG
MAB_4621c|M.abscessus_ATCC_199 ILHYLPLPVP-YTVGDARSFIAASAQEWVAGT--RYGFGFFVMDTGELAG
: : . :. : : . : *
MMAR_4519|M.marinum_M QLTVGNVVHGALRSAWIGYWVASSQTGGGIATGAVALGLDHCFGPVGLHR
MUL_4691|M.ulcerans_Agy99 QLTVGNVVHGALRSAWIGYWVASSQTGGGIATGAVALGLDHCFGPVGLHR
MLBr_00184|M.leprae_Br4923 QLTIGNVTHGALRSAWIGYWVSSSATGGGVATGALALGLDHCFGPVMLHR
MSMEG_5221|M.smegmatis_MC2_155 SLVLFDFRS---DRAEVGFWLTAQSRGRGAAVKALAASSQIVAT-MGLEL
TH_1242|M.thermoresistible__bu SIVLFDIHD---GRAEVGFWLTPQARGRGAARNALRAVVDLAAR-SGLTR
MAB_4621c|M.abscessus_ATCC_199 TCSLKPVTS---GVLEVGYWSAAQHRRKGLTTEAVQRLCQWGFDVLDAHR
: . :*:* :.. * : *: :
MMAR_4519|M.marinum_M VEATVRPENAASRAVLAKVGFREEGLLRRYLEVDRAWRDHLLVGLTVEEV
MUL_4691|M.ulcerans_Agy99 VEATVRPENAASRAVLAKVGFREEGLLRRYLEVDRAWRDHLLVGLTVEEV
MLBr_00184|M.leprae_Br4923 VEATVRPANVASRAVLAKVGFREEGLLRRYLEVDRAWRDHLLMALTIEEV
MSMEG_5221|M.smegmatis_MC2_155 LTARTSPQNLASQRVLERAGFQQKGMPTEQQTPSGVRVPVLTFERPVVEF
TH_1242|M.thermoresistible__bu LDARTHPANEGSQGVLHGAGFVHTRGPEEQVAPSGELVPVLTFERALTAR
MAB_4621c|M.abscessus_ATCC_199 IEWWAVVGNEGSRAVAEAAGFEMEGLLRQRACRRQEPADWWVGGLLPRPE
: . * .*: * .** .
MMAR_4519|M.marinum_M EGSVVSTLVRAGQASRL
MUL_4691|M.ulcerans_Agy99 EGSVVSTLVRAGQASRL
MLBr_00184|M.leprae_Br4923 SGSVTSNLVRAGRASWL
MSMEG_5221|M.smegmatis_MC2_155 RFNV-------------
TH_1242|M.thermoresistible__bu -----------------
MAB_4621c|M.abscessus_ATCC_199 -----------------