For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRYAAFLRGVNVGGINIKMAELARVLTDAGFDDVRTILASGNVLLDSASDADGVRETAERALREAFGYE AWVLVYDLGQLARISAAYPFEREVEDHHSYVTFVSDPAVLDELAALPPGPGEQVRRGDGVLYWQVPRTAT LDSAIGKTMGKKRYKSSTTTRNLRTLDKVLR
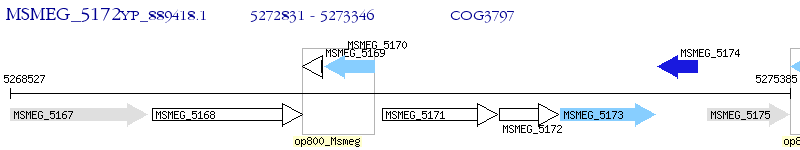
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5172 | - | - | 100% (171) | hypothetical protein MSMEG_5172 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1185c | - | 8e-61 | 67.05% (176) | hypothetical protein Mb1185c |
| M. gilvum PYR-GCK | Mflv_1050 | - | 3e-14 | 32.18% (174) | hypothetical protein Mflv_1050 |
| M. tuberculosis H37Rv | Rv1154c | - | 2e-60 | 66.48% (176) | hypothetical protein Rv1154c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1293c | - | 1e-55 | 63.01% (173) | hypothetical protein MAB_1293c |
| M. marinum M | MMAR_4299 | - | 1e-66 | 70.76% (171) | hypothetical protein MMAR_4299 |
| M. avium 104 | MAV_1291 | - | 4e-65 | 71.10% (173) | hypothetical protein MAV_1291 |
| M. thermoresistible (build 8) | TH_0459 | - | 4e-66 | 70.11% (174) | HYPOTHETICAL PROTEIN Rv1154c |
| M. ulcerans Agy99 | MUL_0995 | - | 4e-66 | 70.18% (171) | hypothetical protein MUL_0995 |
| M. vanbaalenii PYR-1 | Mvan_5780 | - | 4e-13 | 32.22% (180) | hypothetical protein Mvan_5780 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5172|M.smegmatis_MC2_155 -------------------------------------MTRYAAFLRGVNV
TH_0459|M.thermoresistible__bu ----------------------------------MVFVARYVAFLRGVNV
Mb1185c|M.bovis_AF2122/97 MEFPLITANSLSSKTWRAMPRAYVAVASFSGGLVQSGMAKFAAFLRGVNV
Rv1154c|M.tuberculosis_H37Rv MEFPLITANSLSSKTWRAMPRAYVAVASFSGGLVQSGMAKFAAFLRGVNV
MAV_1291|M.avium_104 ----MVVTG----------P-----------GWWNRGMTEYAAFLRGVNV
MMAR_4299|M.marinum_M -------------------------------------MTQFAAFLRGVNV
MUL_0995|M.ulcerans_Agy99 ------MRGSLT-------------------AWCNRIMTQFAAFLRGVNV
MAB_1293c|M.abscessus_ATCC_199 -------------------------------------MTKYAALLRGVNV
Mflv_1050|M.gilvum_PYR-GCK -----------------------------------MGSTRYAALLRGINV
Mvan_5780|M.vanbaalenii_PYR-1 -----------------------------------MGSSRYVALLRGINV
:.:.*:***:**
MSMEG_5172|M.smegmatis_MC2_155 GGIN-IKMA----ELARVLTDAGFDDVRTILASGNVLLDSASDADGVRET
TH_0459|M.thermoresistible__bu GGVN-LKMASGDDSVAAVFTAAGFSAVRTVLATGNVILDSAARPDTVRAT
Mb1185c|M.bovis_AF2122/97 GGVN-LKMA----EVATALTDAGFCNVRTILASGNVLLESTCGAAEVREK
Rv1154c|M.tuberculosis_H37Rv GGVN-LKMA----EVATALTDAGFCNVRTILASGNVLLESTCGAAEVREK
MAV_1291|M.avium_104 GGVN-LKMA----DVKKALTDAGFTAVRTVLASGNVLLQSKAGAAAVRAK
MMAR_4299|M.marinum_M GGVN-LKMA----EVTAALTDAGFTNVRTILASGNVLLEAPGDATTVRQS
MUL_0995|M.ulcerans_Agy99 GGVN-LKMA----EVTAALTDAGFTNLRTILASGNVLLEAPGDATTVRQS
MAB_1293c|M.abscessus_ATCC_199 GGIT-MKMA----DVRDALETAGFAGVTTVLASGNVLLDADEPAATVKER
Mflv_1050|M.gilvum_PYR-GCK GGRNKITMA----DLRAAFSEAGFGGVSTYIQSGNVAFESGRRRASLESD
Mvan_5780|M.vanbaalenii_PYR-1 GGRNKISMP----DLRAVLEDAGFGNVSTYIQSGNVAFESDGARASLESD
** . :.*. .: .: *** : * : :*** ::: :.
MSMEG_5172|M.smegmatis_MC2_155 AERALREAFGYEAWVLVYDLGQLARISAAYP--FEREVEDHHSYVTFVSD
TH_0459|M.thermoresistible__bu AERALRDRFGYDAWVLVYPFDDVAAISAAFP--FERVIPDHHSYVTFVSD
Mb1185c|M.bovis_AF2122/97 TEATLRERFGYDAWALIYDVDTVRTIVAAYP--FECELEGYQSYVTFVAD
Rv1154c|M.tuberculosis_H37Rv TEATLRERFGYDAWALIYDVDTVRTIVTAYP--FECELEGYQSYVTFVAD
MAV_1291|M.avium_104 AEAALRERFGYDAWVLVYDLDTVRRVVDGYP--FEREVDGYQSYVTFVAD
MMAR_4299|M.marinum_M AESTLRNKFGYDAWVLVYDIETVRTINEAYP--FEREVDGLQSYVTFVSD
MUL_0995|M.ulcerans_Agy99 AESTLRNKFGYNAWVLVYDIETVRTINEAYP--FEREVDGLQSYVTFVSD
MAB_1293c|M.abscessus_ATCC_199 LQQVLGERFGYEAWVLVYDLDTIARIIAAFP--WEPEIEGVHSYVMFCSD
Mflv_1050|M.gilvum_PYR-GCK IETLLADRLQSPPVVVVRSHAEMRAIVEEAPADFVARTVDHRRDVVFLKE
Mvan_5780|M.vanbaalenii_PYR-1 IEALLVRLLESPPVVVVRSHRQLRGVVEDAPGEFAERHTEHHRDVIFVKA
: * : . .:: : : * : : * *
MSMEG_5172|M.smegmatis_MC2_155 PAVLDELAALP--PGPGEQVRRGD---GVLYWQVPRTATLDSAIGKTMGK
TH_0459|M.thermoresistible__bu PQVLDELAALD--PGPGEQIAPGD---RVVYWQVRRTQTLTSAMGKTMGK
Mb1185c|M.bovis_AF2122/97 AAILDELSALADTAGPDENISRGPDPLGVLYWQVPKGSTLDSTIGQTMGK
Rv1154c|M.tuberculosis_H37Rv AAILDELSALADTAGPDENISRGPDPLGVLYWQVPKGSTLDSTIGQTMGK
MAV_1291|M.avium_104 AAVLDELAALP--AGPDERIRRGPDPLGVLYWQVPKGSTLDSTIGKTMGK
MMAR_4299|M.marinum_M DSVLDELAALT--AGANEKIHRGD---GVIYWQVPKGATLDSTIGRTMGK
MUL_0995|M.ulcerans_Agy99 DSVLDELAALT--TGANEKIHRGD---GVIYWQVPKGATLDSTIGRTMGK
MAB_1293c|M.abscessus_ATCC_199 PVVLTELAALESELDESEQIAAGD---GVLYWQVPKSQTLGSPVGKTTGK
Mflv_1050|M.gilvum_PYR-GCK PLTADRALEVIQVRDGVDEVWPGP---GVVYFTRVSAERTRSMMNRIVGK
Mvan_5780|M.vanbaalenii_PYR-1 PLTVERAMEVVQLRDGVDDAWPGP---GVVYFTRVIAEKARSRMSRIVGT
. : . : * *:*: * :.: *.
MSMEG_5172|M.smegmatis_MC2_155 KRYKSSTTTRNLRTLDKVLR------
TH_0459|M.thermoresistible__bu KRYKSSTTTRNLRTLERIVG------
Mb1185c|M.bovis_AF2122/97 KRYKSSTTTRNLRTLAKVLR------
Rv1154c|M.tuberculosis_H37Rv KRYKSSTTTRNLRTLAKVLR------
MAV_1291|M.avium_104 PRYKSSTTTRNLRTLVKVLS------
MMAR_4299|M.marinum_M KRYKASTTTRNLRTLDKVLR------
MUL_0995|M.ulcerans_Agy99 KRYKASTTTRNLRTLDKVLR------
MAB_1293c|M.abscessus_ATCC_199 KRYKSTTTTRNLRTLHKLVR------
Mflv_1050|M.gilvum_PYR-GCK PEYR-LMTIRNWATTSQMLSILDGMG
Mvan_5780|M.vanbaalenii_PYR-1 PEYR-LMTIRNWATTTRMLTLLDEMR
.*: * ** * :::