For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAVLSNDQVDAALPNLPGWERAAGALRRSVKFPTFLDGIDAVRRVAEFAEEKDHHPDIDIRWRTVTFALV THAAGGITEKDVQMAEEINRILSD
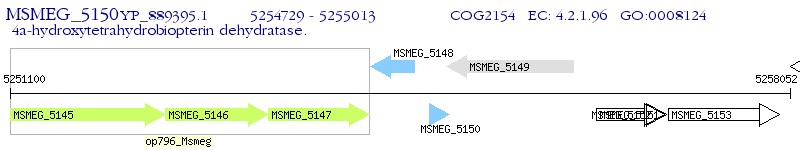
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5150 | phhB | - | 100% (94) | pterin-4-alpha-carbinolamine dehydratase |
| M. smegmatis MC2 155 | MSMEG_2620 | - | 7e-06 | 31.91% (94) | hypothetical protein MSMEG_2620 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1191c | phhB | 2e-35 | 75.28% (89) | pterin-4-alpha-carbinolamine dehydratase |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv3110 | moaB1 | 1e-07 | 32.58% (89) | pterin-4-alpha-carbinolamine dehydratase MoaB1 |
| M. leprae Br4923 | MLBr_01503A | phhB | 4e-32 | 67.02% (94) | pterin-4-alpha-carbinolamine dehydratase |
| M. abscessus ATCC 19977 | MAB_1307c | phhB | 2e-31 | 66.29% (89) | pterin-4-alpha-carbinolamine dehydratase |
| M. marinum M | MMAR_4291 | phhB | 3e-38 | 75.53% (94) | hypothetical protein MMAR_4291 |
| M. avium 104 | MAV_1299 | phhB | 4e-39 | 78.26% (92) | pterin-4-alpha-carbinolamine dehydratase |
| M. thermoresistible (build 8) | TH_0646 | - | 5e-38 | 77.17% (92) | HYPOTHETICAL PROTEIN Rv1159A |
| M. ulcerans Agy99 | MUL_1007 | phhB | 4e-37 | 75.00% (92) | pterin-4-alpha-carbinolamine dehydratase |
| M. vanbaalenii PYR-1 | Mvan_4549 | phhB | 6e-39 | 81.52% (92) | pterin-4-alpha-carbinolamine dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1191c|M.bovis_AF2122/97 ----------------------------MAVLTDEQVDAALHDLNGWQRA
MLBr_01503A|M.leprae_Br4923 ----------------------------MTVLTDEQVDAALLDLNDWKHT
MMAR_4291|M.marinum_M -------------------MNPRRTVGHMAVLTDEQVDAELPGLNGWQRA
MUL_1007|M.ulcerans_Agy99 ------------------------------MLTDEQVDAELPGLNGWQRA
MAV_1299|M.avium_104 ----------------------------MAVLTDEQIDAALPDLDGWERA
Mvan_4549|M.vanbaalenii_PYR-1 ----------------------------MAVLTDEQVDAALPDLNGWERA
MSMEG_5150|M.smegmatis_MC2_155 ----------------------------MAVLSNDQVDAALPNLPGWERA
TH_0646|M.thermoresistible__bu ----------------------------MAVLTDEQVDAALSGLNGWERK
MAB_1307c|M.abscessus_ATCC_199 ----------------------------MALLTDDQINAALAGLPGWTRE
Rv3110|M.tuberculosis_H37Rv MTVSTPEQHEQRASHDASEGKHNVCQGRLAALADAAVSEKLGALPGWQLL
*:: :. * * .*
Mb1191c|M.bovis_AF2122/97 GGVLRRSIKFPTFMAGIDAVRRVAERAEEVNHHPDIDIR-WRTVTFALVT
MLBr_01503A|M.leprae_Br4923 DGALCRSIKFPTFLAGIDAVCRVAEHAETKDHHPDIDIR-YRTVTFILVT
MMAR_4291|M.marinum_M DGALRRSIKFATFLDGIDAVRRVAEHAESKDHHPDIDIR-WRTVTFGLVT
MUL_1007|M.ulcerans_Agy99 DGALRRSIKFATFLDAIDAVRRVAEHAESKDHHPDIDIR-WRTVTFALVT
MAV_1299|M.avium_104 DGALRRSIKFPSFLDGIDAVRRVAEHAESKDHHPDIDIR-WRTVTFALVT
Mvan_4549|M.vanbaalenii_PYR-1 DGALRRSVKFSAFLDGIDAVRRVAEHAEAKDHHPDIDIR-WRTVTFALVT
MSMEG_5150|M.smegmatis_MC2_155 AGALRRSVKFPTFLDGIDAVRRVAEFAEEKDHHPDIDIR-WRTVTFALVT
TH_0646|M.thermoresistible__bu DGALRRSVTFPTFLDGIEAVRRVAERAEQKDHHPDIDIR-WRTVTFALVT
MAB_1307c|M.abscessus_ATCC_199 GDSLRRAVTFDAFLDGIDAVRRIAEHAESVDHHPDIDIR-WRTVTFVLST
Rv3110|M.tuberculosis_H37Rv DMRLSRAFQCTNFDQSIDFMNRVASIANDINHHPDIAVLDKRSVRVTAWT
* *:. * .*: : *:*. *: :***** : *:* . *
Mb1191c|M.bovis_AF2122/97 HAVGGITENDIAMAHDIDAMFGA--------
MLBr_01503A|M.leprae_Br4923 HYADGITKKDITMARDIDRLIGD--------
MMAR_4291|M.marinum_M HSAGGITQNDIDMARDINGIVGD--------
MUL_1007|M.ulcerans_Agy99 HSAGGITQNDIDMARDINGIVGD--------
MAV_1299|M.avium_104 HSEGGITQNDVDMARDINGIVGS--------
Mvan_4549|M.vanbaalenii_PYR-1 HSEGGITDKDVQMARDIDGILG---------
MSMEG_5150|M.smegmatis_MC2_155 HAAGGITEKDVQMAEEINRILSD--------
TH_0646|M.thermoresistible__bu HSAGGITEKDLELAKEIDELIDR--------
MAB_1307c|M.abscessus_ATCC_199 HSEGGITDKDLALAETIDTQIGR--------
Rv3110|M.tuberculosis_H37Rv RKLGYLTDIDFDLAASVEAMYATEFADRPAR
: . :*. *. :* ::