For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVLTDEQVDAALLDLNDWKHTDGALCRSIKFPTFLAGIDAVCRVAEHAETKDHHPDIDIRYRTVTFILV THYADGITKKDITMARDIDRLIGD
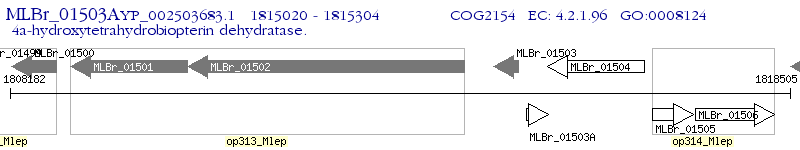
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01503A | phhB | - | 100% (94) | pterin-4-alpha-carbinolamine dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1191c | phhB | 7e-33 | 70.97% (93) | pterin-4-alpha-carbinolamine dehydratase |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1307c | phhB | 6e-26 | 56.99% (93) | pterin-4-alpha-carbinolamine dehydratase |
| M. marinum M | MMAR_4291 | phhB | 1e-34 | 74.47% (94) | hypothetical protein MMAR_4291 |
| M. avium 104 | MAV_1299 | phhB | 7e-35 | 72.04% (93) | pterin-4-alpha-carbinolamine dehydratase |
| M. smegmatis MC2 155 | MSMEG_5150 | phhB | 2e-31 | 67.02% (94) | pterin-4-alpha-carbinolamine dehydratase |
| M. thermoresistible (build 8) | TH_0646 | - | 3e-33 | 71.74% (92) | HYPOTHETICAL PROTEIN Rv1159A |
| M. ulcerans Agy99 | MUL_1007 | phhB | 7e-34 | 73.91% (92) | pterin-4-alpha-carbinolamine dehydratase |
| M. vanbaalenii PYR-1 | Mvan_4549 | phhB | 5e-35 | 74.19% (93) | pterin-4-alpha-carbinolamine dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01503A|M.leprae_Br4923 ---------MTVLTDEQVDAALLDLNDWKHTDGALCRSIKFPTFLAGIDA
Mb1191c|M.bovis_AF2122/97 ---------MAVLTDEQVDAALHDLNGWQRAGGVLRRSIKFPTFMAGIDA
MMAR_4291|M.marinum_M MNPRRTVGHMAVLTDEQVDAELPGLNGWQRADGALRRSIKFATFLDGIDA
MUL_1007|M.ulcerans_Agy99 -----------MLTDEQVDAELPGLNGWQRADGALRRSIKFATFLDAIDA
MAV_1299|M.avium_104 ---------MAVLTDEQIDAALPDLDGWERADGALRRSIKFPSFLDGIDA
Mvan_4549|M.vanbaalenii_PYR-1 ---------MAVLTDEQVDAALPDLNGWERADGALRRSVKFSAFLDGIDA
MSMEG_5150|M.smegmatis_MC2_155 ---------MAVLSNDQVDAALPNLPGWERAAGALRRSVKFPTFLDGIDA
TH_0646|M.thermoresistible__bu ---------MAVLTDEQVDAALSGLNGWERKDGALRRSVTFPTFLDGIEA
MAB_1307c|M.abscessus_ATCC_199 ---------MALLTDDQINAALAGLPGWTREGDSLRRAVTFDAFLDGIDA
:*:::*::* * .* .* : . * *::.* :*: .*:*
MLBr_01503A|M.leprae_Br4923 VCRVAEHAETKDHHPDIDIRYRTVTFILVTHYADGITKKDITMARDIDRL
Mb1191c|M.bovis_AF2122/97 VRRVAERAEEVNHHPDIDIRWRTVTFALVTHAVGGITENDIAMAHDIDAM
MMAR_4291|M.marinum_M VRRVAEHAESKDHHPDIDIRWRTVTFGLVTHSAGGITQNDIDMARDINGI
MUL_1007|M.ulcerans_Agy99 VRRVAEHAESKDHHPDIDIRWRTVTFALVTHSAGGITQNDIDMARDINGI
MAV_1299|M.avium_104 VRRVAEHAESKDHHPDIDIRWRTVTFALVTHSEGGITQNDVDMARDINGI
Mvan_4549|M.vanbaalenii_PYR-1 VRRVAEHAEAKDHHPDIDIRWRTVTFALVTHSEGGITDKDVQMARDIDGI
MSMEG_5150|M.smegmatis_MC2_155 VRRVAEFAEEKDHHPDIDIRWRTVTFALVTHAAGGITEKDVQMAEEINRI
TH_0646|M.thermoresistible__bu VRRVAERAEQKDHHPDIDIRWRTVTFALVTHSAGGITEKDLELAKEIDEL
MAB_1307c|M.abscessus_ATCC_199 VRRIAEHAESVDHHPDIDIRWRTVTFVLSTHSEGGITDKDLALAETIDTQ
* *:** ** :********:***** * ** .***.:*: :*. *:
MLBr_01503A|M.leprae_Br4923 IGD
Mb1191c|M.bovis_AF2122/97 FGA
MMAR_4291|M.marinum_M VGD
MUL_1007|M.ulcerans_Agy99 VGD
MAV_1299|M.avium_104 VGS
Mvan_4549|M.vanbaalenii_PYR-1 LG-
MSMEG_5150|M.smegmatis_MC2_155 LSD
TH_0646|M.thermoresistible__bu IDR
MAB_1307c|M.abscessus_ATCC_199 IGR
..