For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RRVDESRLSWSYSGVALNSEHGADLGVPLVPPPAVTPSPAALRRVLRRARDGVALNVDEAAIAMTARGDD LADLCASAARVRDAGLTTAGRRGPNGRLPVSYSRKVFIPVTHLCRDTCHYCTFVTVPGKLRAQGMGMYME PDEILDVARQGAELGCKEALFTLGDRPEDRWEEARQWLDERGYDSTLDYVRAMAIRVLEETGLLPHLNPG VMSWSELSRLKPVAPSMGMMLETTSRRLFETKGEAHYGSPDKDPAVRLRTLDDAGRLSIPFTTGLLVGIG ETLAERAETMHAIRRSHKEFGHVQEVIVQNFRAKDRTAMASTPDADIDDFLATVAVTRLVLGPKMRIQAP PNLVSRDECLALIGAGVDDWGGVSPLTPDHVNPERPWPALDELADVTAEAGYDLVQRLTAQPQYVQAGAA WIDPRVRAHVDALADPDTGYALDVNPSGLPWQEPDEASESLGRTDLHTAIDAEGRLTDTRSDLDSAFGDW ESIREKVSELAARAPERIDTDVLSALRSAERNPGGCSDDEYLALATADGPALEAVTALADSLRRDAVGDD VTFVVNRNINFTNICYTGCRFCAFAQRKGDADAYSLSVDEVADRAWEAHVAGATEVCMQGGIDPELPVTG YADLVRAVKKRVPSMHVHAFSPMEIANGVTRSGMSIREWLTVLREAGLDTIPGTAAEILDDEVRWVLTKG KLPTSMWIEVITTAHEVGLRSSSTMMYGHVDQPRHWVGHLNVIKSIQDRTGGFTEFVPLPFVHQSSPLYL AGGSRPGPTHRDNRAVHALARIMLHGRIDHIQTSWVKLGIERTQVMLRGGANDLGGTLMEETISRMAGSE NGSAKTVEELVAIAEGIGRPARQRTTTYAPLAA*
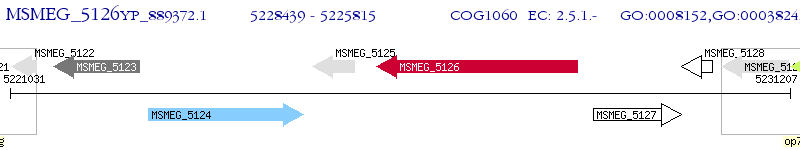
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5126 | fbiC | - | 100% (874) | FO synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1206 | fbiC | 0.0 | 86.88% (846) | FO synthase |
| M. gilvum PYR-GCK | Mflv_2167 | fbiC | 0.0 | 90.92% (859) | FO synthase |
| M. tuberculosis H37Rv | Rv1173 | fbiC | 0.0 | 86.88% (846) | FO synthase |
| M. leprae Br4923 | MLBr_01492 | fbiC | 0.0 | 84.42% (860) | FO synthase |
| M. abscessus ATCC 19977 | MAB_1319 | fbiC | 0.0 | 85.32% (838) | FO synthase |
| M. marinum M | MMAR_4279 | fbiC | 0.0 | 85.93% (846) | F420 biosynthesis protein FbiC |
| M. avium 104 | MAV_1312 | fbiC | 0.0 | 86.07% (854) | FO synthase |
| M. thermoresistible (build 8) | TH_1827 | - | 0.0 | 89.02% (774) | PROBABLE F420 BIOSYNTHESIS PROTEIN FBIC |
| M. ulcerans Agy99 | MUL_1020 | fbiC | 0.0 | 85.46% (846) | FO synthase |
| M. vanbaalenii PYR-1 | Mvan_4533 | fbiC | 0.0 | 91.39% (859) | FO synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2167|M.gilvum_PYR-GCK ---------------MALN-IQHGADLPSPVVPPKS------AAPLPTAL
Mvan_4533|M.vanbaalenii_PYR-1 ---------------MALN-TQRGADLPSPVVPPRS------GAPNPAAL
MSMEG_5126|M.smegmatis_MC2_155 MRRVDESRLSWSYSGVALN-SEHGADLGVPLVPPPA------VTPSPAAL
MMAR_4279|M.marinum_M ---------------MPLTSRQEPTALPSPVVP---------PKPSASAL
MUL_1020|M.ulcerans_Agy99 ---------------MPLTSRQEPTALPSPVVP---------PKPSASAL
MAV_1312|M.avium_104 ---------------MLLTPGEEPTALPDPVVPARASSRAT-PKATASAL
Mb1206|M.bovis_AF2122/97 ---------------MPQPVGRKSTALPSPVVP---------PQANASAL
Rv1173|M.tuberculosis_H37Rv ---------------MPQPVGRKSTALPSPVVP---------PQANASAL
MLBr_01492|M.leprae_Br4923 --------MWGSYTKVSLIESQEPIALSRPVVP---------PKPNTSAL
TH_1827|M.thermoresistible__bu --------------------------------------------------
MAB_1319|M.abscessus_ATCC_1997 --MMNPRVVGFSVRADDLRVTDGCTNVPDDVTPLPNPAFPSRKAASSTAL
Mflv_2167|M.gilvum_PYR-GCK RRVLRRARDGVALNVDEAAIALTARGDDLADLCASAARVRDAGLEAAGRR
Mvan_4533|M.vanbaalenii_PYR-1 RRVLRRARDGVALNVDEAAIALTARGDDLVDLCASAARVRDAGLEAAGRR
MSMEG_5126|M.smegmatis_MC2_155 RRVLRRARDGVALNVDEAAIAMTARGDDLADLCASAARVRDAGLTTAGRR
MMAR_4279|M.marinum_M RRVLRRARDGVALNIDEAAVAMSARGDQLAELCASAARVRDAGLESAGRR
MUL_1020|M.ulcerans_Agy99 RRVLRRARDGVALNIDEAAVAMSARGDQLAELCASAARVRDAGLESAGRH
MAV_1312|M.avium_104 RRVLRRARDGVALNIDEAAVAMTARGDDLADLCASAARVRDAGLESAGRR
Mb1206|M.bovis_AF2122/97 RRVLRRARDGVTLNVDEAAIAMTARGDELADLCASAARVRDAGLVSAGRH
Rv1173|M.tuberculosis_H37Rv RRVLRRARDGVTLNVDEAAIAMTARGDELADLCASAARVRDAGLVSAGRH
MLBr_01492|M.leprae_Br4923 RRVLRRARDGFALNIDEAVVAMTARGEDLADLCASAARVRDVGLETAGRR
TH_1827|M.thermoresistible__bu --------------------------------------------------
MAB_1319|M.abscessus_ATCC_1997 RRVLRRARDGVTLNVDEAALALTARGDDLADLMASAARVRDAGLESGGRL
Mflv_2167|M.gilvum_PYR-GCK GAAGRLPVSYSRKVFIPVTHLCRDKCHYCTFVTVPGKLRAEGKGMFMEPD
Mvan_4533|M.vanbaalenii_PYR-1 GDTGRLPVSYSRKVFIPVTHLCRDTCHYCTFVTVPGRLRAEGKGMFMEPD
MSMEG_5126|M.smegmatis_MC2_155 GPNGRLPVSYSRKVFIPVTHLCRDTCHYCTFVTVPGKLRAQGMGMYMEPD
MMAR_4279|M.marinum_M GTGGGLPITYSRKVFIPVTHLCRDNCHYCTFVTVPGKLRAQGAGMYLEPD
MUL_1020|M.ulcerans_Agy99 GTGGGLPITYSRKVFIPVTHLCRDNCHYCTFVTVPGKLRAQGAGMYLEPD
MAV_1312|M.avium_104 GPGGGLPITYSRKVFIPVTHLCRDNCHYCTFVTVPGKLRARGAGLYLEPD
Mb1206|M.bovis_AF2122/97 GPSGRLAISYSRKVFIPVTRLCRDNCHYCTFVTVPGKLRAQGSSTYMEPD
Rv1173|M.tuberculosis_H37Rv GPSGRLAISYSRKVFIPVTRLCRDNCHYCTFVTVPGKLRAQGSSTYMEPD
MLBr_01492|M.leprae_Br4923 GADGRLPITYSRKVFIPVTHLCRDSCHYCTFVTAPDMLRTQGAGMYLEPN
TH_1827|M.thermoresistible__bu -------VTYSRKVFIPVTHLCRDTCHYCTFVTVPGKLRAAGRGMYLEPD
MAB_1319|M.abscessus_ATCC_1997 GAEGRLPISYSRKVFIPVTHLCRDKCHYCTFVTVPGKLRAQGQGMYMEPD
::**********:****.********.*. **: * . ::**:
Mflv_2167|M.gilvum_PYR-GCK EILDVARRGAELGCKEALFTLGDRPEARWDEARQWLDERGYDSTLDYVRA
Mvan_4533|M.vanbaalenii_PYR-1 EILDVARGGAEMGCKEALFTLGDRPEARWPEARRWLDERGYDSTLDYVRA
MSMEG_5126|M.smegmatis_MC2_155 EILDVARQGAELGCKEALFTLGDRPEDRWEEARQWLDERGYDSTLDYVRA
MMAR_4279|M.marinum_M EILDIARRGAQLGCKEALFTLGDRPEQRWPEAKEWLSERGYDTTLAYVRA
MUL_1020|M.ulcerans_Agy99 EILDIARRGAQLGCKEALFTLGDRPEQRWPEAKEWLSERGYDTTLAYVRA
MAV_1312|M.avium_104 EILDIARRGGELGCKEALFTLGDRPEDRWPEAREWLEARGYDSTLSYVRA
Mb1206|M.bovis_AF2122/97 EILDVARRGAEFGCKEALFTLGDRPEARWRQAREWLGERGYDSTLSYVRA
Rv1173|M.tuberculosis_H37Rv EILDVARRGAEFGCKEALFTLGDRPEARWRQAREWLGERGYDSTLSYVRA
MLBr_01492|M.leprae_Br4923 EILNLARRGSELGCKEALFTLGDRPEDRWAQARDWLAERGYDSTLSYLRA
TH_1827|M.thermoresistible__bu EIVEIARRGAELGCKEALFTLGDRPESRWPEARQWLDERGYDSTLAYVRA
MAB_1319|M.abscessus_ATCC_1997 EILDVARRGAELGCKEALFTLGDRPEERWPEAKQWLDERGYDTTLDYVRA
**:::** *.::************** ** :*: ** ****:** *:**
Mflv_2167|M.gilvum_PYR-GCK MAIRVLEETGLLPHLNPGVMSWSELSRLKPVAPSMGMMLETTSRRLFETR
Mvan_4533|M.vanbaalenii_PYR-1 MAIRVLEETGLLPHLNPGVMSWSELSRLKPVAPSMGMMLETTSRRLFETK
MSMEG_5126|M.smegmatis_MC2_155 MAIRVLEETGLLPHLNPGVMSWSELSRLKPVAPSMGMMLETTSRRLFETK
MMAR_4279|M.marinum_M MAIRVLEETGLLPHLNPGVMSWSEMSLLKPVAPSMGMMLETTSRRLFETK
MUL_1020|M.ulcerans_Agy99 MAIRVLEETGLLPHLNPGVMSWSEMSLLKPVAPSMGMMLETTSRRLFETK
MAV_1312|M.avium_104 MAIRVLEETGLLPHLNPGVMSWSEMARLKPVAPSMGMMLETTSRRLFETK
Mb1206|M.bovis_AF2122/97 MAIRVLEQTGLLPHLNPGVMSWSEMSRLKPVAPSMGMMLETTSRRLFETK
Rv1173|M.tuberculosis_H37Rv MAIRVLEQTGLLPHLNPGVMSWSEMSRLKPVAPSMGMMLETTSRRLFETK
MLBr_01492|M.leprae_Br4923 MAIRVLEETGLLPHLNPGVMSWSELSRLKPVAPSMGMMLETTSRRLFETK
TH_1827|M.thermoresistible__bu MAIRVLEETGLLPHLNPGVMSWSELSLLKPVAPSMGMMLETTSRRLFETK
MAB_1319|M.abscessus_ATCC_1997 MAIRVLEETGLLPHLNPGVMSWAELARLKPVAPSMGMMLETTSRRLFEDR
*******:**************:*:: ********************* :
Mflv_2167|M.gilvum_PYR-GCK GLAHYGSPDKDPAVRLRTLDDAGRLSIPFTTGLLVGIGETLAERAETVHA
Mvan_4533|M.vanbaalenii_PYR-1 GLAHYGSPDKDPAVRLRTLDDAGRLSIPFTTGLLVGIGETLTERAETVHA
MSMEG_5126|M.smegmatis_MC2_155 GEAHYGSPDKDPAVRLRTLDDAGRLSIPFTTGLLVGIGETLAERAETMHA
MMAR_4279|M.marinum_M GLAHYGSPDKDPAVRLRTLTDAGRLSIPFTTGLLVGIGETLAERADTLHA
MUL_1020|M.ulcerans_Agy99 GLAHYGSPDKDPAVRLRTLTDAGRLSIPFTTGLLVGSGETLAERADTLHA
MAV_1312|M.avium_104 GLAHYGSPDKDPAVRLRTLTDAGRLSIPFTTGLLVGIGETLAERADTLHA
Mb1206|M.bovis_AF2122/97 GLAHYGSPDKDPAVRLRVLTDAGRLSIPFTTGLLVGIGETLSERADTLHA
Rv1173|M.tuberculosis_H37Rv GLAHYGSPDKDPAVRLRVLTDAGRLSIPFTTGLLVGIGETLSERADTLHA
MLBr_01492|M.leprae_Br4923 GLAHYGSLDKDPTVRLRVLTDAGRLSIPFTTGLLVGIGETLAERADTLHE
TH_1827|M.thermoresistible__bu GMAHYGSPDKDPAVRLRTLDDAGRLSIPFTTGLLVGIGETVRERAETLHA
MAB_1319|M.abscessus_ATCC_1997 GEAHYGSPDKDPAVRLRTLTDAGRLSIPFTTGLLVGIGENLTERAETIHA
* ***** ****:****.* **************** **.: ***:*:*
Mflv_2167|M.gilvum_PYR-GCK IRRSHKEFGHVQEVIVQNFRAKDHTVMATAPDAGFEDFLATIAVTRLVMG
Mvan_4533|M.vanbaalenii_PYR-1 IRRSHKEFGHVQEVIVQNFRAKDHTAMASVPDAGLDDFLATIAVTRLVMG
MSMEG_5126|M.smegmatis_MC2_155 IRRSHKEFGHVQEVIVQNFRAKDRTAMASTPDADIDDFLATVAVTRLVLG
MMAR_4279|M.marinum_M IRKLHKEFGHVQEVIVQNFRAKEHTAMAAVPDAGIEDYLATVAVARLVLG
MUL_1020|M.ulcerans_Agy99 IRKLHKEFGHVQEVIVQSFRAKEHTAMAAVPDAGIEDYLATVAVARLVLG
MAV_1312|M.avium_104 IRKSHKEFGHVQEVIVQNFRAKEHTAMAAVPDAGIEDYLATVAVARLVLG
Mb1206|M.bovis_AF2122/97 IRKSHKEFGHIQEVIVQNFRAKEHTAMAAFPDAGIEDYLATVAVARLVLG
Rv1173|M.tuberculosis_H37Rv IRKSHKEFGHIQEVIVQNFRAKEHTAMAAFPDAGIEDYLATVAVARLVLG
MLBr_01492|M.leprae_Br4923 IRKSNKEFGHVQEVIVQNFRAKEHTAMAAVPDARIEDYLATVAVARLVLG
TH_1827|M.thermoresistible__bu IRGSHKEFGHVQEVIVQNFRAKQHTAMAGAPDAGFEEFVATIATARLVLG
MAB_1319|M.abscessus_ATCC_1997 IRKVHKEFGHVQEVIVQNFRAKSDTAMKSAPDADIDDFLATIAVTRLVLG
** :*****:******.****. *.* *** :::::**:*.:***:*
Mflv_2167|M.gilvum_PYR-GCK PKVRIQAPPNLVSRQECLALIGAGVDDWGGVSPLTPDHVNPERPWPALDE
Mvan_4533|M.vanbaalenii_PYR-1 PKVRIQAPPNLVSRQECLALIAAGVDDWGGVSPLTPDHVNPERPWPALDE
MSMEG_5126|M.smegmatis_MC2_155 PKMRIQAPPNLVSRDECLALIGAGVDDWGGVSPLTPDHVNPERPWPALDE
MMAR_4279|M.marinum_M PGMRIQAPPNLVSQDECAALIGAGVDDWGGVSPLTPDHVNPERPWPALDD
MUL_1020|M.ulcerans_Agy99 PGMRIQAPPNLVSQDECVALIGAGVDDWGGVSPLTPDHVNPERPWPALDD
MAV_1312|M.avium_104 PGMRIQAPPNLVSRDECRALIAAGIDDWGGVSPLTPDHVNPERPWPALDE
Mb1206|M.bovis_AF2122/97 PGMRIQAPPNLVSGDECRALVGAGVDDWGGVSPLTPDHVNPERPWPALDE
Rv1173|M.tuberculosis_H37Rv PGMRIQAPPNLVSGDECRALVGAGVDDWGGVSPLTPDHVNPERPWPALDE
MLBr_01492|M.leprae_Br4923 PAMRIQAPPNLVSREECLALVTAGVDDWGGVSPLTPDHVNPERPWPALHE
TH_1827|M.thermoresistible__bu PKMRIQAPPNLVSRSECLALLGAGVDDWGGVSPLTPDHVNPERPWPALDE
MAB_1319|M.abscessus_ATCC_1997 PKMRVQAPPNLVSRSECLALIGAGVDDWGGVSPLTPDHVNPERPWPALDD
* :*:******** .** **: **:***********************.:
Mflv_2167|M.gilvum_PYR-GCK LAEVTAEAGYDLVQRLTAQPQYVQAGAAWIDPRVMGHVTALADPESGLAL
Mvan_4533|M.vanbaalenii_PYR-1 LAEVTLEAGYDLVQRLTAQPQYVQAGAAWIDPRVHGHVAALADPDTGYAR
MSMEG_5126|M.smegmatis_MC2_155 LADVTAEAGYDLVQRLTAQPQYVQAGAAWIDPRVRAHVDALADPDTGYAL
MMAR_4279|M.marinum_M LAAVTSGAGYELVQRLTAQPKYVQAGGAWIDPRVRRHVVALADPATGFAR
MUL_1020|M.ulcerans_Agy99 LAAVTSGAGYELVQRLTAQPKYVQAGGAWIDPRVRRHVVALADPATGFAR
MAV_1312|M.avium_104 LAAVTAESGYELVQRLTAQPKYVQAGAAWIDPRVSGHVRALADPVTGLAR
Mb1206|M.bovis_AF2122/97 LAAVTAEAGYDMVQRLTAQPKYVQAGAAWIDPRVRGHVVALADPATGLAR
Rv1173|M.tuberculosis_H37Rv LAAVTAEAGYDMVQRLTAQPKYVQAGAAWIDPRVRGHVVALADPATGLAR
MLBr_01492|M.leprae_Br4923 LAAVTAEAGYTLVQRLTAQPKYVQAGAAWIDPRVRGHVVALADPVTGLAR
TH_1827|M.thermoresistible__bu LAAVTAEAGFELTERLTAQPAYVQAGVAWIDPRVRAHVDALADPATGLAR
MAB_1319|M.abscessus_ATCC_1997 LASVTAEAGYDLVQRLTAQPQYVQAGAAWIDPRVRGHVEALANPETGYAL
** ** :*: :.:****** ***** ******* ** ***:* :* *
Mflv_2167|M.gilvum_PYR-GCK DVNPVGRPWQEPDEATESLGRTDLHAAIDSEGRLTETRSDLGSAFGDWES
Mvan_4533|M.vanbaalenii_PYR-1 DVNPVGRPWQEPDEATESLGRVDLHTAIDTEGRLTTTRSDLGDAFGDWES
MSMEG_5126|M.smegmatis_MC2_155 DVNPSGLPWQEPDEASESLGRTDLHTAIDAEGRLTDTRSDLDSAFGDWES
MMAR_4279|M.marinum_M EVNPVGLPWQEPDDVA-SSGRVDLNAAIDTEGRCTEARSDLDSAFGDWDA
MUL_1020|M.ulcerans_Agy99 EVNPVGLPWQEPDDVA-SSGRVDLNAAIDTEGRCTEARSDLDSAFGDWDA
MAV_1312|M.avium_104 DVNPVGLPWQEPDDVG-SSGRVDLNAAIDTQGRNSETRSDIDSAFGDWES
Mb1206|M.bovis_AF2122/97 DVNPVGMPWQEPDDVA-SWGRVDLGAAIDTQGRNTAVRSDLASAFGDWES
Rv1173|M.tuberculosis_H37Rv DVNPVGMPWQEPDDVA-SWGRVDLGAAIDTQGRNTAVRSDLASAFGDWES
MLBr_01492|M.leprae_Br4923 DVNPVGMPWQEPDDVE-SAGRMDINTAIDTEGRNTEARSDLDSAFGDWES
TH_1827|M.thermoresistible__bu DVRPVGRPWQEPDEAAGSLGRVDLHTAIDTEGRHTRTRSDLGEAFGDWDS
MAB_1319|M.abscessus_ATCC_1997 DISPTGLPWQEPDETWESTGRVDLHAAIDSEGRNTDTRSDLASAFGDWES
:: * * ******:. * ** *: :***::** : .***: .*****::
Mflv_2167|M.gilvum_PYR-GCK IRAKVEELAAR---APERIDTDVLAALRAAERDPAGCSDDEYLALATADG
Mvan_4533|M.vanbaalenii_PYR-1 IRAKVEELAAR---APERIDTDVLAALRSAERDPAGCSDDEYLALATADG
MSMEG_5126|M.smegmatis_MC2_155 IREKVSELAAR---APERIDTDVLSALRSAERNPGGCSDDEYLALATADG
MMAR_4279|M.marinum_M VRSHVHELAAR---GPERIDTDVLAALRSAERSPAGCSDEEYLALATADG
MUL_1020|M.ulcerans_Agy99 VRSHVHDLAAR---NPERIDTDVLAALRSAERSPAGCSDEEYLALATADG
MAV_1312|M.avium_104 IRAHVHELAAQGARAPERLDSDVLAALRSAERDPAGCTDSEYLSLATADG
Mb1206|M.bovis_AF2122/97 IREQVHELAVR---APERIDTDVLAALRSAERAPAGCTDGEYLALATADG
Rv1173|M.tuberculosis_H37Rv IREQVHELAVR---APERIDTDVLAALRSAERAPAGCTDGEYLALATADG
MLBr_01492|M.leprae_Br4923 IRAHVHELADC---APERIDTDVLAALRSAERDPAGCTDDEYLALATADG
TH_1827|M.thermoresistible__bu IRAKISELTAR---APERIDTDVLAALRSAERDPAGCTDDEYLALATADG
MAB_1319|M.abscessus_ATCC_1997 IREHVRELNAR---APEKVGADVLAALRSAERDPAGCTDDEYLALATAEG
:* :: :* **::.:***:***:*** *.**:* ***:****:*
Mflv_2167|M.gilvum_PYR-GCK PALEAVTALADSLRRDVVGDDVTFVVNRNINFTNICYTGCRFCAFAQRKG
Mvan_4533|M.vanbaalenii_PYR-1 PALDAVTALADSLRRDTVGDDVTFVVNRNINFTNICYTGCRFCAFAQRKG
MSMEG_5126|M.smegmatis_MC2_155 PALEAVTALADSLRRDAVGDDVTFVVNRNINFTNICYTGCRFCAFAQRKG
MMAR_4279|M.marinum_M PALEAVAALADSLRRDAVGDDVTFVVNRNINFTNVCYTGCRFCAFAQRKG
MUL_1020|M.ulcerans_Agy99 PALEAVAALADSLRRDAVGDDVTFVVNRNINFTNVCYTGCRFCAFAQRKG
MAV_1312|M.avium_104 PALEAVAALADSLRRDTVGDDVTFVVNRNINFTNICYTGCRFCAFAQRKG
Mb1206|M.bovis_AF2122/97 PALEAVAALADSLRRDVVGDEVTFVVNRNINFTNICYTGCRFCAFAQRKG
Rv1173|M.tuberculosis_H37Rv PALEAVAALADSLRRDVVGDEVTFVVNRNINFTNICYTGCRFCAFAQRKG
MLBr_01492|M.leprae_Br4923 PALEAVTALADSLRRDVVGDDVTFVVNRNINFTNICYTGCRFCAFAQRKG
TH_1827|M.thermoresistible__bu PALDAVAALADSLRRDTVGDDVTYVVNRNINFTNICYTGCRFCAFAQRKG
MAB_1319|M.abscessus_ATCC_1997 PAMDALAALADSIRADVVGDDVTYVVNRNINFTNICYTGCRFCAFAQRKG
**::*::*****:* *.***:**:**********:***************
Mflv_2167|M.gilvum_PYR-GCK DADAFSLSTSEVADRAWEAHVAGATEVCMQGGIDPELPVTGYADLVRAVK
Mvan_4533|M.vanbaalenii_PYR-1 DADAYSLSTAEVADRAWEAHVAGATEVCMQGGIDPELPVTGYADLVRAVK
MSMEG_5126|M.smegmatis_MC2_155 DADAYSLSVDEVADRAWEAHVAGATEVCMQGGIDPELPVTGYADLVRAVK
MMAR_4279|M.marinum_M DADAYSLSADEVADRAWEAHVAGATEVCMQGGIDPELPVTGYADLVRAVK
MUL_1020|M.ulcerans_Agy99 DADAYSLSADEVADRAWEAHVAGATEVCMQGGIDPELPVTGYADLVRAVK
MAV_1312|M.avium_104 DADAYSLSTEEVADRAWEAHVAGATEVCMQGGIDPELPVTGYADLVRAVK
Mb1206|M.bovis_AF2122/97 DADAYSLSVGEVADRAWEAHVAGATEVCMQGGIDPELPVTGYADLVRAVK
Rv1173|M.tuberculosis_H37Rv DADAYSLSVGEVADRAWEAHVAGATEVCMQGGIDPELPVTGYADLVRAVK
MLBr_01492|M.leprae_Br4923 DTDAYSLSREEVAERAWEAHVQGATEVCMQGGIDPELPVTGYVDLVRAVK
TH_1827|M.thermoresistible__bu DADAYSLSVEEVADRAWEAHVAGATEVCMQGGIDPELPVTGYADLVRAVK
MAB_1319|M.abscessus_ATCC_1997 DADAFSLSAEEVGDRAWEAYVAGATEVCMQGGIDPELPVTGYADLVRAVK
*:**:*** **.:*****:* ********************.*******
Mflv_2167|M.gilvum_PYR-GCK ARVPSMHVHAFSPMEIANGVTRSGTSIREWLTALREAGLDTIPGTAAEIL
Mvan_4533|M.vanbaalenii_PYR-1 ARVPSMHVHAFSPMEIANGVTRSGTSIREWLTALREAGLDTIPGTAAEIL
MSMEG_5126|M.smegmatis_MC2_155 KRVPSMHVHAFSPMEIANGVTRSGMSIREWLTVLREAGLDTIPGTAAEIL
MMAR_4279|M.marinum_M ARVPSMHVHAFSPMEIANGVTKSGLSIREWLTALREAGLGTIPGTAAEIL
MUL_1020|M.ulcerans_Agy99 ARVPSMHVHAFSPMEIANGVTKSGLSIREWLTALREAGLGTIPGTAAEIL
MAV_1312|M.avium_104 ARVPSMHVHAFSPMEIANGVTKSGLSIREWLISLREAGLDTIPGTAAEIL
Mb1206|M.bovis_AF2122/97 ARVPSMHVHAFSPMEIANGVTKSGLSIREWLIGLREAGLDTIPGTAAEIL
Rv1173|M.tuberculosis_H37Rv ARVPSMHVHAFSPMEIANGVTKSGLSIREWLIGLREAGLDTIPGTAAEIL
MLBr_01492|M.leprae_Br4923 TRVPSMHVHAFSPMEIANGVAKSGFSIREWLISLREAGLDTIPGTAAEIL
TH_1827|M.thermoresistible__bu ARVPSMHVHAFSPMEIANGVTRSGLSIREWLISLREAGLDTIPGTAAEIL
MAB_1319|M.abscessus_ATCC_1997 KRVPSMHVHAFSPMEIVNGASKGGQSVRDWLTELRAAGLDTIPGTAAEIL
***************.**.::.* *:*:** ** ***.**********
Mflv_2167|M.gilvum_PYR-GCK DDEVRWVLTKGKLPTSMWIEVVTTAHEVGLRSSSTMMYGHVDTPRHWVGH
Mvan_4533|M.vanbaalenii_PYR-1 DDEVRWVLTKGKLPTSMWIEVVTTAHEVGLRSSSTMMYGHVDTPRHWVGH
MSMEG_5126|M.smegmatis_MC2_155 DDEVRWVLTKGKLPTSMWIEVITTAHEVGLRSSSTMMYGHVDQPRHWVGH
MMAR_4279|M.marinum_M DDEVRWVLTKGKLPTSLWIEIVSTAHEVGLRSSSTMMYGHVDSPRHWIAH
MUL_1020|M.ulcerans_Agy99 DDEVRWVLTKGKLPTSLWIEIVSTAHEVGLRSSSTMMYGHVDSPRHWIAH
MAV_1312|M.avium_104 DDEVRWVLTKGKLPTSLWIEIVTTAHEVGLRSSSTMMYGHVDNPRHWVGH
Mb1206|M.bovis_AF2122/97 DDEVRWVLTKGKLPTSLWIEIVTTAHEVGLRSSSTMMYGHVDSPRHWVAH
Rv1173|M.tuberculosis_H37Rv DDEVRWVLTKGKLPTSLWIEIVTTAHEVGLRSSSTMMYGHVDSPRHWVAH
MLBr_01492|M.leprae_Br4923 DDEVRWVLTKGKLPTSMWIEIVTTAHEVGLRSSSTMMYGHVDGPRHWVAH
TH_1827|M.thermoresistible__bu DDEVRWVLTKGKLPTSMWIEIVTTAHEVGLRSSSTMMYGHVDTPRHWIGH
MAB_1319|M.abscessus_ATCC_1997 DDEIRWVLTKGKLPASEWIDVISTAHEVGLRSSSTMMYGHVDTPKHWVAH
***:**********:* **::::******************* *:**:.*
Mflv_2167|M.gilvum_PYR-GCK LNVLRDIQDRTGGFTEFVPLPFVHQSSPLYLAGGARPGPTHRDNRAVHAL
Mvan_4533|M.vanbaalenii_PYR-1 LNVLRGIQDRTGGFTEFVPLPFVHQSSPLYLAGGSRPGPTHRDNRAVHAL
MSMEG_5126|M.smegmatis_MC2_155 LNVIKSIQDRTGGFTEFVPLPFVHQSSPLYLAGGSRPGPTHRDNRAVHAL
MMAR_4279|M.marinum_M LNVLRDIQDRTGGFTEFVPLPFVHQNSPLYLAGAARPGPTHRDNRAVHAL
MUL_1020|M.ulcerans_Agy99 LNVLRDIQDRTGGFTEFVPLPFVHQNSPLYLAGAARPGPTHRDNRAVHAL
MAV_1312|M.avium_104 LNVLRDIQDRTGGFTEFVPLPFVHQSSPLYLAGAARPGPTHRDNRAVHAL
Mb1206|M.bovis_AF2122/97 LNVLRDIQDRTGGFTEFVPLPFVHQNSPLYLAGAARPGPSHRDNRAVHAL
Rv1173|M.tuberculosis_H37Rv LNVLRDIQDRTGGFTEFVPLPFVHQNSPLYLAGAARPGPSHRDNRAVHAL
MLBr_01492|M.leprae_Br4923 LQVLRDIQDRTGGFTEFVPLPFVHQNSPLYLAGAARPGPTHRDNRAVHAL
TH_1827|M.thermoresistible__bu LRILRDIQDRTGGFTEFVPLPFVHQSSPLYLAGGARPGPTHRDNRAVHAL
MAB_1319|M.abscessus_ATCC_1997 LRVLAGIQDRTGGFTEFVPLPFVHQSAPLYLAGAARPGPTNRDNRAVHAL
*.:: .*******************.:******.:****::*********
Mflv_2167|M.gilvum_PYR-GCK ARIMLHGRISNIQTSWVKLGIERTQVMLTGGANDLGGTLMEETISRMAGS
Mvan_4533|M.vanbaalenii_PYR-1 ARIMLHGRISHIQTSWVKLGTERTQVMLRGGANDLGGTLMEETISRMAGS
MSMEG_5126|M.smegmatis_MC2_155 ARIMLHGRIDHIQTSWVKLGIERTQVMLRGGANDLGGTLMEETISRMAGS
MMAR_4279|M.marinum_M ARIMLHGRIPHIQTSWVKLGVERTRVMLSGGANDLGGTLMEETISRMAGS
MUL_1020|M.ulcerans_Agy99 ARIMLHGRIPHIQTSWVKLGVERTRVMLSGGANDLGGTLMEETISRMAGS
MAV_1312|M.avium_104 ARIMLHGRISQIQTSWVKLGVQRTQVMLNGGANDLGGTLMEETISRMAGS
Mb1206|M.bovis_AF2122/97 ARIMLHGRISHIQTSWVKLGVRRTQVMLEGGANDLGGTLMEETISRMAGS
Rv1173|M.tuberculosis_H37Rv ARIMLHGRISHIQTSWVKLGVRRTQVMLEGGANDLGGTLMEETISRMAGS
MLBr_01492|M.leprae_Br4923 ARIMLHGRISHIQTSWVKLGVERTQAMLNGGANDLGGTLMEETISRMAGS
TH_1827|M.thermoresistible__bu ARIMLHGRINHIQTSWVKLGVERTQVMLQGGANDLGGTLMEETISRMAGS
MAB_1319|M.abscessus_ATCC_1997 ARIMLHGRIDNIQTSWVKLGIEGTRVMLQSGANDLGGTLMEETISRMAGS
********* :********* . *:.** .********************
Mflv_2167|M.gilvum_PYR-GCK ENGSAKSVAELVAIAEGIGRPARQRSTDYSPLAA--
Mvan_4533|M.vanbaalenii_PYR-1 ENGSAKTVAELVAIAEGIGRPARQRTTDYTPLAA--
MSMEG_5126|M.smegmatis_MC2_155 ENGSAKTVEELVAIAEGIGRPARQRTTTYAPLAA--
MMAR_4279|M.marinum_M EHGSAKTAAELAAIAAGIGRPARQRTTTYASLAA--
MUL_1020|M.ulcerans_Agy99 EHGSAKTAAELAAIAAGIGRPARQRTTTYASLAA--
MAV_1312|M.avium_104 EHGSAKTVAELTAIAEGIGRPARQRTTDYTPLAA--
Mb1206|M.bovis_AF2122/97 EHGSAKTVAELVAIAEGIGRPARQRTTTYALLAA--
Rv1173|M.tuberculosis_H37Rv EHGSAKTVAELVAIAEGIGRPARQRTTTYALLAA--
MLBr_01492|M.leprae_Br4923 EYGSAKTVAELIAIAEGIGRTARQRTTTYALRGA--
TH_1827|M.thermoresistible__bu EHGSAKTVAELVAIAEGIGRPARQRTTTYTPLAA--
MAB_1319|M.abscessus_ATCC_1997 EHGSAKTIAELEEIADGIGRPAVERTTTYARRPSAA
* ****: ** ** ****.* :*:* *: :