For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKPISGKTTSILRRLAAAACVGAVTMAMTSCTSDERDIPSAGGTAEVGATNDINPQDVANLRQGGNLRLA LPGFPSNFNTLHIDGNEASLGGLMRATLPRAYRVGPDGSTTVNTDYFTSVELTSTDPQVVTYTINPEAVW TDGTPITWEDIAAQIHATSGKDKAFQIASLNGSDRVASVTRGVDDRQAVVTFAEPFSEWQGMFAGNTMLL PKSSTSDPEVFNKGQLNGPGPSAGPFMVSNVDRGAQRITLTRNPKWWGEPPVLDSITYTVLDDAARIPAL QNNALDATGVASLDELEIARRTPGISIRRAPSPSWYHFTFNGAPGSILADKALRQAVAKGIDRNIITTVA QRGLADDPKPLNNHIFVAGQEGYQDNSEVVAFDPEKAKAELDALGWTLKPGAQFREKDGKRLTIRDVLYD SQSTRQIATIAQNSLAQIGVELRIEAKPGSGFFTDWIIPGNFDIAQFSWVGDAFSLCCLNQIYTTDADSN FGRISSPEVDTKAIATMGELDPEKARTMANELDKLIWEEVFSLPLFQSPGNVAVRSNLANYGPAGIGDID ITKVGFMK
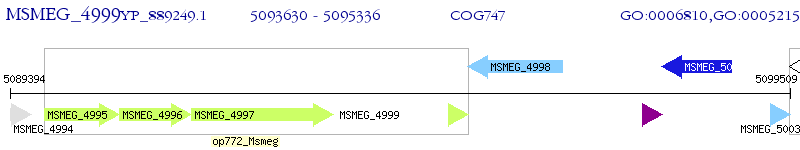
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4999 | - | - | 100% (568) | extracellular solute-binding protein, family protein 5 |
| M. smegmatis MC2 155 | MSMEG_5130 | - | 2e-18 | 23.36% (488) | extracellular solute-binding protein, family protein 5 |
| M. smegmatis MC2 155 | MSMEG_4101 | - | 2e-17 | 25.80% (438) | ABC-type transporter, substrate binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1311c | oppA | 0.0 | 68.25% (548) | periplasmic oligopeptide-binding lipoprotein OppA |
| M. gilvum PYR-GCK | Mflv_2258 | - | 0.0 | 67.75% (555) | extracellular solute-binding protein |
| M. tuberculosis H37Rv | Rv1280c | oppA | 0.0 | 68.25% (548) | periplasmic oligopeptide-binding lipoprotein OppA |
| M. leprae Br4923 | MLBr_01121 | - | 0.0 | 64.70% (558) | putative extracellular solute-binding dependent transport |
| M. abscessus ATCC 19977 | MAB_2878c | - | 1e-19 | 23.37% (505) | hypothetical protein MAB_2878c |
| M. marinum M | MMAR_4139 | oppA | 0.0 | 64.31% (552) | periplasmic oligopeptide-binding lipoprotein OppA |
| M. avium 104 | MAV_1429 | - | 0.0 | 66.61% (551) | extracellular solute-binding protein, family protein 5 |
| M. thermoresistible (build 8) | TH_0742 | oppA | 0.0 | 73.39% (545) | PROBABLE PERIPLASMIC OLIGOPEPTIDE-BINDING LIPOPROTEIN OPPA |
| M. ulcerans Agy99 | MUL_4002 | oppA | 0.0 | 63.85% (545) | periplasmic oligopeptide-binding lipoprotein OppA |
| M. vanbaalenii PYR-1 | Mvan_4433 | - | 0.0 | 68.83% (555) | extracellular solute-binding protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2258|M.gilvum_PYR-GCK ---------------------------MTVFKGAMLRLFAAGSVAALVLA
Mvan_4433|M.vanbaalenii_PYR-1 -----------------------------------MRLLAVGSVVALLVS
MSMEG_4999|M.smegmatis_MC2_155 ---------------------MKPISGKTTSILRRLAAAACVGAVTMAMT
TH_0742|M.thermoresistible__bu ------------------------------------------MVVVLALA
Mb1311c|M.bovis_AF2122/97 MADRGQRRGCAPGIASALRASFQGKSRPWTQTRYWAFALLTPLVVAMVLT
Rv1280c|M.tuberculosis_H37Rv MADRGQRRGCAPGIASALRASFQGKSRPWTQTRYWAFALLTPLVVAMVLT
MMAR_4139|M.marinum_M MLTNADR---HDGVQPGPGARSRRPTRPWAR-RRIRCAGAALLTVTLLLA
MUL_4002|M.ulcerans_Agy99 -----------------------------------------MLTVTLLLA
MLBr_01121|M.leprae_Br4923 -------------------------------MIMLSRIVAALLVVGLILT
MAV_1429|M.avium_104 ------------------------------MGRCGFRPAALVAAVVLAVA
MAB_2878c|M.abscessus_ATCC_199 ---------------------------------MDSRSPARTGARRIARI
. :
Mflv_2258|M.gilvum_PYR-GCK GCSSGEQSVPAAGGNS---ELGATADINPQDPATLRQGGNLRLALTGFPT
Mvan_4433|M.vanbaalenii_PYR-1 GCSSGQNDVPSTGGSA---ELGATADINPQDPATLQQGGNLRLALTGFPS
MSMEG_4999|M.smegmatis_MC2_155 SCTSDERDIPSAGGTA---EVGATNDINPQDVANLRQGGNLRLALPGFPS
TH_0742|M.thermoresistible__bu GCAREDTDTPAAGGPA---ELGTTNDINPQDPADLEQGGNLRLALTALPP
Mb1311c|M.bovis_AF2122/97 GCSASGTQLELAPTADRRAAVGTTSDINQQDPATLQDGGNLRLSLTDFPP
Rv1280c|M.tuberculosis_H37Rv GCSASGTQLELAPTADRRAAVGTTSDINQQDPATLQDGGNLRLSLTDFPP
MMAR_4139|M.marinum_M GCSAS-TQLPAATSGS--TAVGSSSDINPQDPATLEDGGDLRLSLTDFPP
MUL_4002|M.ulcerans_Agy99 GCSAS-TQLPAATSGS--TAVGSSSDINPQDPATLEDGGDLRLSLTDFPP
MLBr_01121|M.leprae_Br4923 GCSASNRLVPSAASNA---ALGKTSDINPQDPATLKDGGNLRLALTDFPA
MAV_1429|M.avium_104 GCSPAINLVPETGQNAR---IGTTSDINPRDPATLQDGGNLRLALTDFPP
MAB_2878c|M.abscessus_ATCC_199 GAIVTTVLLAAGLLTS----------------CWTSAAKTLDYAVDGRLT
.. . . * :: .
Mflv_2258|M.gilvum_PYR-GCK NFNNLHIDGNLGEIGRMYRPTLPRAFFIKPDGEMTVNSDYFTSVELTSTE
Mvan_4433|M.vanbaalenii_PYR-1 NFNNLHIDGNLGEIGRMYRPTLPRAFFIKPDGEMTVNSDYFTDVELTSTD
MSMEG_4999|M.smegmatis_MC2_155 NFNTLHIDGNEASLGGLMRATLPRAYRVGPDGSTTVNTDYFTSVELTSTD
TH_0742|M.thermoresistible__bu NFNPLHIDGNVADAAGMLKATLPRAFRIAPDGTATVNTDYFTSIELTSED
Mb1311c|M.bovis_AF2122/97 NFNILHIDGNNAEVAAMMKATLPRAFIIGPDGSTTVDTNYFTSIELTRTA
Rv1280c|M.tuberculosis_H37Rv NFNILHIDGNNAEVAAMMKATLPRAFIIGPDGSTTVDTNYFTSIELTRTA
MMAR_4139|M.marinum_M NFNILHIDGNSAEVAAMVKATLPRAFIIGPDGSTTVDHDYFTSVELTKTN
MUL_4002|M.ulcerans_Agy99 NFNILHIDGNSAEVAAMVKATLPRAFIIGPDGSTTVDHDYFTSVELTKTN
MLBr_01121|M.leprae_Br4923 NFNILHIDGNNAEVTSMMKATMPRAFIIGTDGSTTVDTNYFTSIKLTSTS
MAV_1429|M.avium_104 NFNILHIDGNSADVSAMMKATLPRAFVIGPDGSTTVDTDYFTSVELTGTA
MAB_2878c|M.abscessus_ATCC_199 TYNVNSVAGNASAGAQAFNRVLTGFGFHGPDGQVIADHD-FGTVEVVGRI
.:* : ** . . .:. .** .: : * :::.
Mflv_2258|M.gilvum_PYR-GCK PQVVTYTINPKATWSNGRPVTWEDLAAQINATSGKDERFLFASPNGSERV
Mvan_4433|M.vanbaalenii_PYR-1 PQVVTYTINPNAVWTNERPVTWEDIAAQINATSGKDKRFLFASPNGSERV
MSMEG_4999|M.smegmatis_MC2_155 PQVVTYTINPEAVWTDGTPITWEDIAAQIHATSGKDKAFQIASLNGSDRV
TH_0742|M.thermoresistible__bu PQVVTYTINPDAVWTDGTPITWEDIAAQIHATSGKDPAFAIASSNGADRV
Mb1311c|M.bovis_AF2122/97 PQVVTYTINPEAVWSDGTPITWRDIASQIHAISGADKAFEIASSSGAERV
Rv1280c|M.tuberculosis_H37Rv PQVVTYTINPEAVWSDGTPITWRDIASQIHAISGADKAFEIASSSGAERV
MMAR_4139|M.marinum_M PQVITYTINPKAFWSDGTPITWRDIASQIHATSGADNRFEIASSGGSDRV
MUL_4002|M.ulcerans_Agy99 PQVITYTINPKAFWSDGTPVTWRDIASQIHATSGADNRFEIASSGGSDRV
MLBr_01121|M.leprae_Br4923 PQVVTYTINPKAVWSDGTPITWRDIDRQIHANSGKNKAFEIAGTNGCERV
MAV_1429|M.avium_104 PQVVTYTINPKAVWSDGTPITWEDIASQIHALSGADKTFEIAGSSGAERV
MAB_2878c|M.abscessus_ATCC_199 PLVLDYTINEKAVYSDGKPVTCEDMVLSWFAQSGRMPDFDTASTAGYADI
* *: **** .* ::: *:* .*: . * ** * *. * :
Mflv_2258|M.gilvum_PYR-GCK ASVTKGVDDRQAVITFASH--YSDWRGMFAGNGLLYPKEMTQDPD-----
Mvan_4433|M.vanbaalenii_PYR-1 ASVTRGVDDRQAVVTFAKH--YADWRGMFAGNGMLYPKEITQDPE-----
MSMEG_4999|M.smegmatis_MC2_155 ASVTRGVDDRQAVVTFAEP--FSEWQGMFAGNTMLLPKSSTSDPE-----
TH_0742|M.thermoresistible__bu ESVTRGVDDRQAVITFAKP--YAEWRGMFSGNGMLLPKSMTATPE-----
Mb1311c|M.bovis_AF2122/97 ASVTRGVDDRQAVVTFAKP--YAEWRGMFAGNGMLLPASMTATPE-----
Rv1280c|M.tuberculosis_H37Rv ASVTRGVDDRQAVVTFAKP--YAEWRGMFAGNGMLLPASMTATPE-----
MMAR_4139|M.marinum_M ASVTPGVDDRQAIVTFAKP--YAEWRGMFAGNGMLLPSSMTATPE-----
MUL_4002|M.ulcerans_Agy99 ASVIPGVDDRQAMVTFAKP--YAEWRGMFAGNGMLLPSSMTATPE-----
MLBr_01121|M.leprae_Br4923 ASIRPGANDRQAIVTFAQP--YAEWRGMFAGNGMLLPATMTATPE-----
MAV_1429|M.avium_104 ASVTRGVDDRQAVVTFAKP--YAEWRGMFAGNGMLLPKSMTATPE-----
MAB_2878c|M.abscessus_ATCC_199 STVDCKPGQKTARVTFAPDRAFTDWTQLFTATTMLPWHVVADKLGGNIDL
:: .:: * :*** :::* :*:.. :* :
Mflv_2258|M.gilvum_PYR-GCK -----------------------AFNKGFLNGPGPSAGPFEITTIDRGAQ
Mvan_4433|M.vanbaalenii_PYR-1 -----------------------AFNKGFLTGPGPSAGPFMITTVDRGAQ
MSMEG_4999|M.smegmatis_MC2_155 -----------------------VFNKGQLNGPGPSAGPFMVSNVDRGAQ
TH_0742|M.thermoresistible__bu -----------------------AFNTAQRDGPGPSAGPFMITSIDRGAQ
Mb1311c|M.bovis_AF2122/97 -----------------------AFNKGQLDGPGPSAGPFVVSALDRTAQ
Rv1280c|M.tuberculosis_H37Rv -----------------------AFNKGQLDGPGPSAGPFVVSALDRTAQ
MMAR_4139|M.marinum_M -----------------------AFNKGQLAGPGPSAGPFIVTSLDRSAQ
MUL_4002|M.ulcerans_Agy99 -----------------------AFNKGQLTGPGPSAGPFIVTSLDRSAQ
MLBr_01121|M.leprae_Br4923 -----------------------AFNKGQLDRPGPSAGPFIVSSLDRTTQ
MAV_1429|M.avium_104 -----------------------AFNKGQLEGPGPSAGPFIVSSLDRTTQ
MAB_2878c|M.abscessus_ATCC_199 VGAIASNDVPALQKIADFWNNQWNLDTDLDLSKFPSSGPYKLDGYNDDGS
::. **:**: : : .
Mflv_2258|M.gilvum_PYR-GCK RIVLTRNPKWWGTPPLLDSMTYTVLDDAAVIPALENNALDVAGLATLDDL
Mvan_4433|M.vanbaalenii_PYR-1 RITLERNPKWWGTPPVLDRITYTVLDDAAMLPALENNALDSIGLGTLDDL
MSMEG_4999|M.smegmatis_MC2_155 RITLTRNPKWWGEPPVLDSITYTVLDDAARIPALQNNALDATGVASLDEL
TH_0742|M.thermoresistible__bu RIVLTRNPDWWGDPPLLDSITYTVLDDAARIPALQNNALDATGLATIDEL
Mb1311c|M.bovis_AF2122/97 RIVLTRNPRWWGARPRLDSITYLVLDDAARLPALQNNTIDATGVGTLDQL
Rv1280c|M.tuberculosis_H37Rv RIVLTRNPRWWGARPRLDSITYLVLDDAARLPALQNNTIDATGVGTLDQL
MMAR_4139|M.marinum_M RIVLTRNPKWWGKKPRLDSITYLVLDDAARIPALQNFTIDATGIGSLDQL
MUL_4002|M.ulcerans_Agy99 RIVLIRNPKWWGKKPRLHSITYLVLDDAARIPALQNFTIDATGIGSLDQL
MLBr_01121|M.leprae_Br4923 RIVLTHNPKWWGTPPRLDSITYLVLDDAARLPALQNNTIDATGVGTLDQL
MAV_1429|M.avium_104 RIVLSRNPKWWGTRPRLDSITYLVLDDAARLPALQNNTIDATGVGTLDQL
MAB_2878c|M.abscessus_ATCC_199 -VVLVANDKWWGNPPVTKRVT-VWPSGIDVQKRLSDGDLEVVDVAAGSAG
:.* * *** * . :* .. *.: :: .:.: .
Mflv_2258|M.gilvum_PYR-GCK ERARRANGVSIRRAPAPNWYHLTFNGAEGALLSDPALRQAISKGIDRQAI
Mvan_4433|M.vanbaalenii_PYR-1 ERARRAQGVTIRRAPAPNWYHLTLNGAEGALLSDPALRAAITKGIDRQAI
MSMEG_4999|M.smegmatis_MC2_155 EIARRTPGISIRRAPSPSWYHFTFNGAPGSILADKALRQAVAKGIDRNII
TH_0742|M.thermoresistible__bu TIAQRTPGISIRRAPGNSWYHFTFNGAPGSILEDKALRQAIAKGIDRQTI
Mb1311c|M.bovis_AF2122/97 TIAARTKGISIRRAPGPSWYHFTLNGAPGSILADKALRLAIAKGIDRYTI
Rv1280c|M.tuberculosis_H37Rv TIAARTKGISIRRAPGPSWYHFTLNGAPGSILADKALRLAIAKGIDRYTI
MMAR_4139|M.marinum_M TIAERTKGISIRRAPAPSWSHFTLNGAPGAVLADPALRLAVCKGIDRRTI
MUL_4002|M.ulcerans_Agy99 TIAERTKGISIRRAPAPSWSHFTLNGAPGAVLADPALRLAVCKGIDRRTI
MLBr_01121|M.leprae_Br4923 TTAQRTNGISIRRTPAPRWSHFTFNGAVGSILADQGLRLAVSKGIDRRTI
MAV_1429|M.avium_104 TIAEHTAGISIRRAPAPAWSHFTFNGAPGSILADRALRVAVSKGIDRQTI
MAB_2878c|M.abscessus_ATCC_199 TLTAP-DGFVRTEEPGSGVEQLIFG--AGGSVGAPDLRRAVALCTPRDAI
: *. . *. :: :. *. : ** *: * *
Mflv_2258|M.gilvum_PYR-GCK AAVSQRGLTDAPVALNNHIYLAGQEGYQDNS--VGFDPEAAKRELDELGW
Mvan_4433|M.vanbaalenii_PYR-1 TAVSQRGLTDDPAALNNHIYLAGQEGYQDNS--IGFDPEAAKRELDALGW
MSMEG_4999|M.smegmatis_MC2_155 TTVAQRGLADDPKPLNNHIFVAGQEGYQDNSEVVAFDPEKAKAELDALGW
TH_0742|M.thermoresistible__bu ANVTQRGLVDNPVPLNNHIFVRGQEGYQDNSDVVAYNPEQAKAELDALGW
Mb1311c|M.bovis_AF2122/97 ARVAQYGLTSDPVPLNNHVFVAGQDGYQDNSGVVAYNPEQAKRELDALGW
Rv1280c|M.tuberculosis_H37Rv ARVAQYGLTSDPVPLNNHVFVAGQDGYQDNSGVVAYNPEQAKRELDALGW
MMAR_4139|M.marinum_M AKVALYGLTNDPAPLNNHVFVAGQEGYQDNS--SPYDPEQAKRELDALGW
MUL_4002|M.ulcerans_Agy99 AKVALYGLTNDPAPLNNHVFVAGQEGYQDNS--SPYDPEQAKRELDALGW
MLBr_01121|M.leprae_Br4923 AKVVQYGLANNPEPLNNHVYVDGQEGYQDNSGVVGYDPEQAQHELDALGW
MAV_1429|M.avium_104 AKVVQYGLTSNPVALGNHIYVAGQQGYQDNSTTVPYDPAAAGRELDALGW
MAB_2878c|M.abscessus_ATCC_199 AGIAGVPVMNARLDTSIGDSFASVEAAAEAGRFMRSDPIAARAAAANRPL
: : : . . . . :. : . :* *
Mflv_2258|M.gilvum_PYR-GCK TLN--GQFREKDGKRLTIRDVFYDGASTRGIAQIAQNQLTQIGVNLELVP
Mvan_4433|M.vanbaalenii_PYR-1 TLN--GQFREKDGKPLTLRDVFYDGASTRAIAQVAQNQLAQIGVNLELVP
MSMEG_4999|M.smegmatis_MC2_155 TLKPGAQFREKDGKRLTIRDVLYDSQSTRQIATIAQNSLAQIGVELRIEA
TH_0742|M.thermoresistible__bu RLN--GQFREKDGRRLVIRDVLFDSLSTRQFAQIAQSNLAEIGVKLEIDA
Mb1311c|M.bovis_AF2122/97 RRS--GAFREKDGRQLVIRDLFYDAQSTRQFAQIAQHTLAQIGVKLELQA
Rv1280c|M.tuberculosis_H37Rv RRS--GAFREKDGRQLVIRDLFYDAQSTRQFAQIAQHTLAQIGVKLELQA
MMAR_4139|M.marinum_M KLN--GKFREKDGHQLVVRLLFYEAQSSRQFAQVAQYSLAQIGVKLELQS
MUL_4002|M.ulcerans_Agy99 KLN--GKFREKDGHQLVVRLLFYEAQSSRQFAQVAQYSLAQIGVKLELQS
MLBr_01121|M.leprae_Br4923 KLN--GQFREKDGRQLVIRYLFYDAQGSREFAQLAQHSLAQIGVKLELQA
MAV_1429|M.avium_104 KLH--GRFREKDGRQLVIRDLFYDAQGSRQFAQIAQHSLAQIGVRLELVA
MAB_2878c|M.abscessus_ATCC_199 TLR--VGYQAPNPRRAAIVSAMAQACEP---AGITVQDVSSPETGPHSLR
:: : : .: : :. . * :: ::. . .
Mflv_2258|M.gilvum_PYR-GCK SAGGRLFTEFITPGNFDIAQFAWGGDAFPLGGLTQIYASGGESNFGKIGS
Mvan_4433|M.vanbaalenii_PYR-1 AAGGSLFPDYITPGNFDIAQFAWGGDAFPLGGLTQIYASNGESNYGKIGS
MSMEG_4999|M.smegmatis_MC2_155 KPGSGFFTDWIIPGNFDIAQFSWVGDAFSLCCLNQIYTTDADSNFGRISS
TH_0742|M.thermoresistible__bu KGATGFFSDHIIPGNFDIAQFTWVGDAFPLSGLTQIYASYGESNFGKIGS
Mb1311c|M.bovis_AF2122/97 KSGSGFFSDYVNVGAFDIAQFGWVGDAFPLSSLTQIYASDGESNFGKIGS
Rv1280c|M.tuberculosis_H37Rv KSGSGFFSDYVNVGAFDIAQFGWVGDAFPLSSLTQIYASDGESNFGKIGS
MMAR_4139|M.marinum_M RSGSGFFSNYVNVGAFDIALFGWVGDAFPLSSLTQIYMSDGESNFGKIGS
MUL_4002|M.ulcerans_Agy99 RSGSGFFSNYVNVGAFDIALFGWVGDAFPLSSLTQIYMSDGESNFGKIGS
MLBr_01121|M.leprae_Br4923 KAGSGFFTNYVNVGAFDIVQFGWVGDAFPLSALTQIYQSDGESNFGKISS
MAV_1429|M.avium_104 RAGSGFFTNYINVGAFDMAQFGWLGDAFPLSALTQIYKSDGESNFGKIGS
MAB_2878c|M.abscessus_ATCC_199 DNQVDALLSSIGGVPGSGSTGSWLMDAYALR-------SREGKNPANYAN
: . : . * **:.* : .* .. ..
Mflv_2258|M.gilvum_PYR-GCK PQIDAKIEETLSELDPAKARTSANELDRMIWEVGHSLPLFQAP-GNVAVR
Mvan_4433|M.vanbaalenii_PYR-1 PQVDAKIEETLSELDPAKARTLANELDKMIWEIGHSLPLFQAP-GNVAVR
MSMEG_4999|M.smegmatis_MC2_155 PEVDTKAIATMGELDPEKARTMANELDKLIWEEVFSLPLFQSP-GNVAVR
TH_0742|M.thermoresistible__bu PEIDAKIEETLSELDPERARELANEIDQMLFEEVFSLPLTQSP-GNVAVR
Mb1311c|M.bovis_AF2122/97 PQIDAAIERTLAELDPGKARALANQVDELIWAEGFSLPLTQSP-GTVAVR
Rv1280c|M.tuberculosis_H37Rv PQIDAAIERTLAELDPGKARALANQVDELIWAEGFSLPLTQSP-GTVAVR
MMAR_4139|M.marinum_M PQIDAAIERTLQELDPAKALALANEVDKLIWAEGFSLPLTQSP-GTIAVR
MUL_4002|M.ulcerans_Agy99 PQIDAAIERTLQELDPAKALALANQVDKLIWAEGFSLPLTQSP-GTIAVR
MLBr_01121|M.leprae_Br4923 PAVDAAIERTLAELDPAKARALANEVDKLLWAEGFSLPLTQSP-GDVAVR
MAV_1429|M.avium_104 PQIDAAIERTLQELDPDKARALANDLDKLIWAEGFSLPLTQSP-GDVAVR
MAB_2878c|M.abscessus_ATCC_199 GQIDGIIAALAVTTNARDQSRLLGDASAILWNDLPTLPLYRQPRVLIAPK
:* :. .: . ::: :*** : * :* :
Mflv_2258|M.gilvum_PYR-GCK SDLANYGPAGIGDLNFSAIGFTK--
Mvan_4433|M.vanbaalenii_PYR-1 SNLANYGPAGIGDINYSAIGFMK--
MSMEG_4999|M.smegmatis_MC2_155 SNLANYGPAGIGDIDITKVGFMK--
TH_0742|M.thermoresistible__bu SNLANFGAFGLGDADYTKIGFMKP-
Mb1311c|M.bovis_AF2122/97 STLANFGATGLADLDYTAIGFMRR-
Rv1280c|M.tuberculosis_H37Rv STLANFGATGLADLDYTAIGFMRR-
MMAR_4139|M.marinum_M STLANFGARGLADLDYTAIGFMRS-
MUL_4002|M.ulcerans_Agy99 STLANFGARGLADLDYTAIGFMRS-
MLBr_01121|M.leprae_Br4923 STLANFGAAGFGDLNYTAIGFMQN-
MAV_1429|M.avium_104 STLANFGAAGLGDLNYTAIGFTRS-
MAB_2878c|M.abscessus_ATCC_199 SMYAATPSVTRWGAGWNMDRWVLPT
* * . . . . :