For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRREDGRLDDELRPVVITRGFTSHPAGSVLVEFGQTRVMCTASVTEGVPRWRKGSGQGWLTAEYAMLPAA THDRSDRESVKGRIGGRTQEISRLIGRSLRACIDLAALGENTIAIDCDVLQADGGTRTAAITGAYVALSD AVTWLAAAGRLSDPRPLSCAIAAVSVGVVDGRVRVDLPYSEDSRAEVDMNVVATDTGTLVEIQGTGEGAT FPRSTLDKMLDAALGATEQLFVLQREALDAPYPGVLPEGPAPKKAFGS*
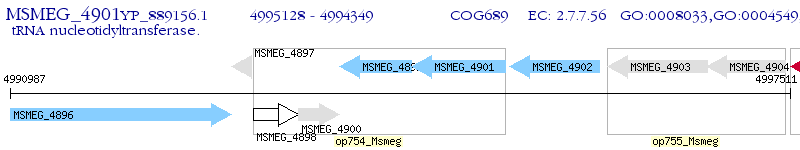
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4901 | rph | - | 100% (259) | ribonuclease PH |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1375 | rph | 1e-126 | 84.94% (259) | ribonuclease PH |
| M. gilvum PYR-GCK | Mflv_2358 | rph | 1e-133 | 90.77% (260) | ribonuclease PH |
| M. tuberculosis H37Rv | Rv1340 | rph | 1e-127 | 85.33% (259) | ribonuclease PH |
| M. leprae Br4923 | MLBr_01174 | rph | 1e-127 | 85.33% (259) | ribonuclease PH |
| M. abscessus ATCC 19977 | MAB_1484 | rph | 1e-129 | 88.42% (259) | ribonuclease PH |
| M. marinum M | MMAR_4042 | rph | 1e-131 | 88.03% (259) | ribonuclease RphA |
| M. avium 104 | MAV_1557 | rph | 1e-131 | 87.26% (259) | ribonuclease PH |
| M. thermoresistible (build 8) | TH_1015 | rphA | 1e-131 | 89.62% (260) | PROBABLE RIBONUCLEASE RPHA (RNase PH) (tRNA |
| M. ulcerans Agy99 | MUL_3908 | rph | 1e-129 | 87.26% (259) | ribonuclease PH |
| M. vanbaalenii PYR-1 | Mvan_4287 | rph | 1e-133 | 90.77% (260) | ribonuclease PH |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2358|M.gilvum_PYR-GCK -----------------MSRREDGRLDDELRPVTITRGFTTHPAGSVLVE
Mvan_4287|M.vanbaalenii_PYR-1 -----------------MSRREDGRLDDELRPVTITRGFTTHPAGSVLVE
TH_1015|M.thermoresistible__bu -----------------VSRREDGRLDDELRPVVITRGYTSHPAGSVLVE
MAB_1484|M.abscessus_ATCC_1997 -----------------MSRREDGRQDDELRPVTITRGFTSHPAGSVLIE
MSMEG_4901|M.smegmatis_MC2_155 -----------------MSRREDGRLDDELRPVVITRGFTSHPAGSVLVE
Mb1375|M.bovis_AF2122/97 -----------------MSKREDGRLDHELRPVIITRGFTENPAGSVLIE
Rv1340|M.tuberculosis_H37Rv -----------------MSKREDGRLDHELRPVIITRGFTENPAGSVLIE
MLBr_01174|M.leprae_Br4923 -----------------MSTRADGRLDDELRAVVITRGFTEHPAGSVLIE
MAV_1557|M.avium_104 -----------------MTRREDGRLDDELRPLVITRGFTEHPAGSVLIE
MMAR_4042|M.marinum_M MSGALGLVSVGRARVGVVSKREDGRRDDELRPVVITRGFTENPAGSVLIE
MUL_3908|M.ulcerans_Agy99 -----------------MSKREDGRRDDELRPVVITRGFTENPAGSVLIE
:: * *** *.***.: ****:* :******:*
Mflv_2358|M.gilvum_PYR-GCK FGETRVMCTASVTEGVPRWRKGSGQGWLTAEYAMLPAATHDRSDRESVKG
Mvan_4287|M.vanbaalenii_PYR-1 FGQTRVMCTASVTEGVPRWRKGSGQGWLTAEYAMLPAATHERSGRESVKG
TH_1015|M.thermoresistible__bu FGHTRVLCTASVLEGVPRWRKGSGQGWLTAEYAMLPAATHERSDRESVKG
MAB_1484|M.abscessus_ATCC_1997 FGQTRVMCTASATEGVPRWRKGSGLGWLTAEYAMLPASTHTRSDRESVKG
MSMEG_4901|M.smegmatis_MC2_155 FGQTRVMCTASVTEGVPRWRKGSGQGWLTAEYAMLPAATHDRSDRESVKG
Mb1375|M.bovis_AF2122/97 FGHTKVLCTASVTEGVPRWRKATGLGWLTAEYAMLPSATHSRSDRESVRG
Rv1340|M.tuberculosis_H37Rv FGHTKVLCTASVTEGVPRWRKATGLGWLTAEYAMLPSATHSRSDRESVRG
MLBr_01174|M.leprae_Br4923 FGHTKVMCTASPTEGVPRWRKGSGKGWLTAEYAMLPSATHTRSERESVKG
MAV_1557|M.avium_104 FGHTKVMCTASVTEGVPRWRKGSGLGWLTAEYAMLPSATHTRSDRESVKG
MMAR_4042|M.marinum_M FGRTKVMCTASVTEGVPRWRKGSGLGWLTAEYAMLPSATHTRSDRESVKG
MUL_3908|M.ulcerans_Agy99 FGRTKVMCTASVTEGVSRWRKGSGLGWLTAEYAMLPSATHTRSDRESVKG
**.*:*:**** ***.****.:* ***********::** ** ****:*
Mflv_2358|M.gilvum_PYR-GCK RVGGRTQEISRLVGRSLRACIDLAALGENTIAIDCDVLQADGGTRTAAIT
Mvan_4287|M.vanbaalenii_PYR-1 RVGGRTQEISRLVGRSLRACIDLAALGENTIAIDCDVLQADGGTRTAAIT
TH_1015|M.thermoresistible__bu RLGGRTQEISRLIGRSLRACIDLNALGENTIAIDCDVLQADGGTRTAAIT
MAB_1484|M.abscessus_ATCC_1997 RVGGRTQEISRLIGRSLRACIDLSALGENTIAIDCDVLQADGGTRTAAIT
MSMEG_4901|M.smegmatis_MC2_155 RIGGRTQEISRLIGRSLRACIDLAALGENTIAIDCDVLQADGGTRTAAIT
Mb1375|M.bovis_AF2122/97 RLSGRTQEISRLISRSLRACIDLAALGENTIAIDCDVLQADGGTRTAAIT
Rv1340|M.tuberculosis_H37Rv RLSGRTQEISRLIGRSLRACIDLAALGENTIAIDCDVLQADGGTRTAAIT
MLBr_01174|M.leprae_Br4923 RLSGRTQEISRLIGRSLRACIDLAALGENTIAVDCDVLQADGGTRTAAVT
MAV_1557|M.avium_104 RLSGRTQEISRLIGRSLRACIDLAALGENTIAVDCDVLQADGGTRTAAIT
MMAR_4042|M.marinum_M RVGGRTQEISRLVGRSLRACIDLAALGENTIAVDCDVLQADGGTRTAAIT
MUL_3908|M.ulcerans_Agy99 RVGGRTQEISRLVGRSLRACIDLAALGENTIAVDCDVLQADGGTRTAAIT
*:.*********:.********* ********:***************:*
Mflv_2358|M.gilvum_PYR-GCK GAFVALSDAVTYLGAAGKLSDPRPLSCAIAAVSVGVVDGRVRVDLPYTED
Mvan_4287|M.vanbaalenii_PYR-1 GAYVALADAVTYLAAAGKLSDPRPLSCAIAAVSVGVVDGRIRVDLPYTED
TH_1015|M.thermoresistible__bu GAYVALADAVTYLAAAEKLSDPWPLSCAIAAISVGVVDGRVRVDLPYSED
MAB_1484|M.abscessus_ATCC_1997 GAYVALADAVTYLAAHGKLADDEPLSCAIAAVSVGVVDGRVRVDLPYEED
MSMEG_4901|M.smegmatis_MC2_155 GAYVALSDAVTWLAAAGRLSDPRPLSCAIAAVSVGVVDGRVRVDLPYSED
Mb1375|M.bovis_AF2122/97 GAYVALADAVTYLSAAGKLSDPRPLSCAIAAVSVGVVDGRIRVDLPYEED
Rv1340|M.tuberculosis_H37Rv GAYVALADAVTYLSAAGKLSDPRPLSCAIAAVSVGVVDGRIRVDLPYEED
MLBr_01174|M.leprae_Br4923 GAYVALADAVTYLSAAGRLLDPRPLSCAIAAVSVGVVDGRIRVDLSYEED
MAV_1557|M.avium_104 GAFVALADAVTYLSAAGKLSDPRPLSCAIAAVSVGVVDGRIRVDLPYEED
MMAR_4042|M.marinum_M GAYVALADAVNYLAAAGKLSDPRPLSCAIAAVSVGVVDGRVRVDLPYEED
MUL_3908|M.ulcerans_Agy99 GAYVALADAVNYLAAAGKLSDPRPLSCAIAAVSVGVVDGRVRVDLPYEED
**:***:***.:*.* :* * ********:********:****.* **
Mflv_2358|M.gilvum_PYR-GCK SRAEVDMNVVATDTGTLVEIQGTGEGATFPRSTLDKMLDAAMAACDQLFE
Mvan_4287|M.vanbaalenii_PYR-1 SRAEVDMNVVATDTGTLVEIQGTGEGATFPRSTLDKMLDAAMAACDQLFE
TH_1015|M.thermoresistible__bu ARAEVDMNVVATDTGTLVEVQGTGEGATFPRSTLDKMLDAALAACDQLFA
MAB_1484|M.abscessus_ATCC_1997 SRAEVDMNVVATDTGTLVEVQGTGEGATFARSTLDKLLDAAVGAADQLFV
MSMEG_4901|M.smegmatis_MC2_155 SRAEVDMNVVATDTGTLVEIQGTGEGATFPRSTLDKMLDAALGATEQLFV
Mb1375|M.bovis_AF2122/97 SRAEVDMNVVATDTGTLVEIQGTGEGATFARSTLDKLLDMALGACDTLFA
Rv1340|M.tuberculosis_H37Rv SRAEVDMNVVATDTGTLVEIQGTGEGATFARSTLDKLLDMALGACDTLFA
MLBr_01174|M.leprae_Br4923 SRAEVDMNVIATDTGTLVEIQGTGEGATFPRSTLDKLLDMALGACDKLFV
MAV_1557|M.avium_104 ARAEVDMNVVATDTGTLVEVQGTGEGATFPRSTLDKLLDAALAACDKLFA
MMAR_4042|M.marinum_M SRAEVDMNVVATDTGTLVEVQGTGEGATFPRSTLDKLLDMALGACDTLFA
MUL_3908|M.ulcerans_Agy99 SRAEVDMNVVATDTGTLVEVQGTGEGATFPRSTLDKLLDMALGACDTLFA
:********:*********:*********.******:** *:.* : **
Mflv_2358|M.gilvum_PYR-GCK IQRAALELPYPGTLPESAEPAKKAFGS
Mvan_4287|M.vanbaalenii_PYR-1 IQRAALELPYPGVLPEPGEPAKKAFGS
TH_1015|M.thermoresistible__bu VQREALELPYPGVLPAPTSPPKKAFGS
MAB_1484|M.abscessus_ATCC_1997 LQKEALALPYPGVLP--DAPQKKAFGS
MSMEG_4901|M.smegmatis_MC2_155 LQREALDAPYPGVLPEGPAP-KKAFGS
Mb1375|M.bovis_AF2122/97 AQRDALALPYPGVLPQGPPP-PKAFGT
Rv1340|M.tuberculosis_H37Rv AQRDALALPYPGVLPQGPPP-PKAFGT
MLBr_01174|M.leprae_Br4923 AQRDALALPYQGMSPEGPPP-PKAFGS
MAV_1557|M.avium_104 AQREALKLPYPGVLPEGPPP-PKVFGS
MMAR_4042|M.marinum_M AQRDALALPYPGVLPEGPPP-SKAFGS
MUL_3908|M.ulcerans_Agy99 AQCDALALPYPGVLPEGPPP-SKAFGS
* ** ** * * * *.**: